* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download WormPset-2015_NoAnswers
X-inactivation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Koinophilia wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Frameshift mutation wikipedia , lookup
Microevolution wikipedia , lookup
Oncogenomics wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
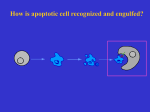
2015 C. elegans problem set I. Basic concepts of diploid genetics and linkage You cross a dpy-5(e61) male (“dumpy” body shape phenotype, recessive mutation) with an unc-13(e51) hermaphrodite (“uncoordinated” movement phenotype, recessive mutation). a. What phenotype(s) do expect to see in F1 cross progeny? Sex ratio? b. You single (i.e. move individual worms onto separate plates and let them self) several of the F1 cross progeny at a stage that you are certain they have not mated with their siblings. What genotypic and phenotypic ratios do you expect to see in the F2 if the two mutations are unlinked? (Draw out the dihybrid cross). How do these ratios change if the mutations are linked? c. At this point, let’s assume that the specific Dpy and Unc genes are linked. From the F2 progeny plate (see above) you single out several Dpy animals, 1 per plate (again, you are certain that they have not mated). What do you expect to see phenotypically in their progeny (F3 relative to the starting cross)? d. Of the 32 Dpy F2 animals that you single, 2 of the worms give rise to progeny (F3) that are ¼ DpyUnc. How many map units are Dpy and Unc apart? 2. Mapping using polymorphisms (SNPs) You have isolated a mutation that appears to be due to inactivation of a single locus. Your screen was started in the N2-Bristol background strain. To identify the mutated gene, you cross your mutant to the CB4856 Hawaiian mapping strain. You pick F1 cross progeny, allow them to self, and from their progeny (F2), you pick animals that once again display the mutant phenotype. You pick 20 such animals. These animals are then placed individually on separate plates and allowed to lay eggs. About a week later, all 20 plates are filled with animals. From each plate you wash some of the animals away for DNA extraction and retain the plates (each plate will still have lots of animals). Using SNP mapping, you generate the following results (a gel showing the SNIP-SNP patterns for each chromosomal marker are shown. Lanes one and two show the band patterns for the N2 and CB strains as controls, and lanes 1-10 show the band patterns for each of 10 F2 clones that are tested): 1 Chrom. I N2 CB _ _ 1 _ _ 2 _ _ 3 _ _ 4 _ _ 5 6 7 8 _ _ _ _ _ _ _ _ _ _ _ _ Chrom II N2 CB _ _ _ 1 _ _ _ 2 4 _ _ _ 5 6 7 8 _ _ 3 _ _ _ _ _ _ _ _ _ _ _ Chrom III N2 CB _ _ 1 _ _ 2 _ 3 _ 4 _ 5 7 _ 8 _ 6 _ _ 1 2 3 4 5 6 8 _ _ _ _ _ _ _ _ _ _ _ _ 7 _ 1 _ 2 _ 3 _ 4 _ 5 _ 6 _ _ Chrom IV N2 CB _ _ _ 9 _ 10 _ _ _ 9 _ 10 _ _ _ 9 _ 10 _ _ _ 9 _ _ _ 10 _ _ _ 7 _ _ 8 _ 9 _ 10 _ 7 8 _ _ _ 9 _ 10 _ _ _ _ Chrom V N2 _ CB _ Chrom X N2 CB _ _ _ 1 _ _ _ 2 _ _ _ 3 _ _ _ 4 _ _ _ 5 _ _ 6 _ _ _ _ _ _ _ a) Which chromosome does this mutation map to? Why? b) What is a rough estimate of map position of your mutation in relation to the SNP on that chromosome? 2 3. Epistasis analysis and pathway building: During programmed cell death cells are killed off and the corpses are engulfed and cleared by neighboring cells. From your first worm discussion paper, you know: ced-9 --| ced-4 ced-3 programmed cell death Once a cell has undergone programmed cell death, its corpse is quickly engulfed and cleared away. Corpse engulfment genes of programmed cell death are identified as mutations in which cell death still occurs leaving a visibly identifiable cell corpse but one that is not cleared away. Consider: Genotype ced-1(lf) ced-3(lf) ced-1(lf), ced-3(lf) phenotype cells die, but leave ugly corpses cells do not die cells do not die a) What is the epistatic relationship of ced-3 and ced-1? Loss of function mutations in ced-1, ced-2, ced-7, and ced-10 were identified as engulfment genes as null mutations in each of these genes left, on average, one corpse per pharynx (C. elegans feeding organ). However, more than one cell undergoes programmed cell death in the pharynx, implying that some corpse engulfment is still occurring in each of the single mutant backgrounds. The following results were obtained from mutant combinations (all cases are null alleles): ced-1; ced-2: average four corpses/pharynx ced-1; ced-7: average one corpse/pharynx ced-1. ced-10: average four corpses/pharynx ced-2; ced-7: average four corpses/pharynx ced-2; ced-10: average one corpse/pharynx ced-7; ced-10 average four corpses/pharynx b) What can you conclude about relationships between these four ced genes? 3