Chapter 14 Guided Reading
... 50. You will need to know the difference between all the gene mutations. Use the diagram below to familiarize yourself with the mutations. ...
... 50. You will need to know the difference between all the gene mutations. Use the diagram below to familiarize yourself with the mutations. ...
ALE 10.
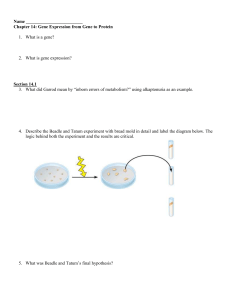
... e.) Translate the mRNA produced above into a peptide. Remember that ribosomes read mRNA in only one specific direction. Use three letter abbreviations to show the amino acid sequence below. 19. List in order the sequence of events that take place when a DNA sequence (i.e. gene) is used to direct the ...
... e.) Translate the mRNA produced above into a peptide. Remember that ribosomes read mRNA in only one specific direction. Use three letter abbreviations to show the amino acid sequence below. 19. List in order the sequence of events that take place when a DNA sequence (i.e. gene) is used to direct the ...
Gene Expression
... 3. An amine group (an electron buzzing around nitrogen. This means it can donate an electron). Amine groups are basic since they remove hydrogen and donate an electron. 4. They all have at least one hydrogen coming off the central carbon. AAs differ in their R group (called the functional group). Th ...
... 3. An amine group (an electron buzzing around nitrogen. This means it can donate an electron). Amine groups are basic since they remove hydrogen and donate an electron. 4. They all have at least one hydrogen coming off the central carbon. AAs differ in their R group (called the functional group). Th ...
Chapter 18: Regulation of Gene Expression
... Feedback inhibition is a recurring mechanism throughout biological systems. In the case of E. coli regulating tryptophan synthesis, is it positive or negative inhibition? Explain your choice. ...
... Feedback inhibition is a recurring mechanism throughout biological systems. In the case of E. coli regulating tryptophan synthesis, is it positive or negative inhibition? Explain your choice. ...
A1991GH39300001
... whids there were a number of published proceIn the nearly 20 years since the procethue was dures. This wasdone just asa control. It worked fine published, it ha been cited hi thousand,of psth&awhen mRNA was extracted from retics, but failed dons and is still the most airiness t.,aj~ij~j~ of when ext ...
... whids there were a number of published proceIn the nearly 20 years since the procethue was dures. This wasdone just asa control. It worked fine published, it ha been cited hi thousand,of psth&awhen mRNA was extracted from retics, but failed dons and is still the most airiness t.,aj~ij~j~ of when ext ...
RNA polymerase
... Eukaryotes multicellular evolved to maintain constant internal conditions while facing changing external conditions ...
... Eukaryotes multicellular evolved to maintain constant internal conditions while facing changing external conditions ...
Dr Ishtiaq Transcription
... Shortly after the discovery of splicing came the realization that the exons in some genes were not utilized in the same way in every cell or stage of development. In other words exons could be skipped or added. This means that variations of a protein (called isoforms) can be produced from the same g ...
... Shortly after the discovery of splicing came the realization that the exons in some genes were not utilized in the same way in every cell or stage of development. In other words exons could be skipped or added. This means that variations of a protein (called isoforms) can be produced from the same g ...
Gene Regulation
... • Near the lac operon is another gene, called lacI, or just “i”. It codes for the lac repressor protein, which plays an essential role in lac operon control. The lac repressor gene is expressed “constitutively”, meaning that it is always on (but at a low level). It is a completely separate gene, pro ...
... • Near the lac operon is another gene, called lacI, or just “i”. It codes for the lac repressor protein, which plays an essential role in lac operon control. The lac repressor gene is expressed “constitutively”, meaning that it is always on (but at a low level). It is a completely separate gene, pro ...
Objectives • Describe the process of DNA transcription. • Explain
... In prokaryotic cells, the mRNA transcribed from a gene directly serves as the messenger molecule that is translated into a protein. But this is not the case in eukaryotic cells. In a eukaryotic cell, the RNA transcribed in the nucleus is modified or processed before it leaves the nucleus as mRNA to ...
... In prokaryotic cells, the mRNA transcribed from a gene directly serves as the messenger molecule that is translated into a protein. But this is not the case in eukaryotic cells. In a eukaryotic cell, the RNA transcribed in the nucleus is modified or processed before it leaves the nucleus as mRNA to ...
... RNA molecule off a DNA template, begins when RNA polymerase attaches to a promoter. Elongation of an RNA molecule occurs through the process of complementary base pairing until there is a DNA stop sequence. Messenger RNA (mRNA), which now carries codons, is processed before it leaves the nucleus; in ...
Lecture 6 The connection between genes, proteins and metabolism
... 3. This definition was modified when it was discovered that many genes code for proteins that are not enzymes e.g. hemoglobin one gene codes for one protein 4. It was modified again when it was discovered that some proteins contain more than one polypeptide chain each of which is encoded by a sepa ...
... 3. This definition was modified when it was discovered that many genes code for proteins that are not enzymes e.g. hemoglobin one gene codes for one protein 4. It was modified again when it was discovered that some proteins contain more than one polypeptide chain each of which is encoded by a sepa ...
ASTR 380 The Origins of Life on Earth
... Cyanobacteria Thylakoid structure is a complex structure which contains at least 335 different proteins… Simpler but not so simple…. ...
... Cyanobacteria Thylakoid structure is a complex structure which contains at least 335 different proteins… Simpler but not so simple…. ...
Gene Expression
... • Variations in snRNPs (as well as other proteins) occur in different cells and recognize slightly different splicing signals. • Some of the splicing proteins also assist in transporting mRNA out of the nucleus. ...
... • Variations in snRNPs (as well as other proteins) occur in different cells and recognize slightly different splicing signals. • Some of the splicing proteins also assist in transporting mRNA out of the nucleus. ...
reading guide
... Feedback inhibition is a recurring mechanism throughout biological systems. In the case of E. coli regulating tryptophan synthesis, is it positive or negative inhibition? Explain your choice. ...
... Feedback inhibition is a recurring mechanism throughout biological systems. In the case of E. coli regulating tryptophan synthesis, is it positive or negative inhibition? Explain your choice. ...
U - Helena High School
... Before making proteins, Your cell must first make RNA • Question: • How does RNA (ribonucleic acid) differ from DNA (deoxyribonucleic acid)? ...
... Before making proteins, Your cell must first make RNA • Question: • How does RNA (ribonucleic acid) differ from DNA (deoxyribonucleic acid)? ...
The Effectiveness of Three input RNA-based Gene
... mRNA transcript and preventing expression of the gene. However, when the desired ligand binds to the aptamer region, a conformation change occurs that disrupts the active conformation of the hammerhead ribozyme. This ligand-bound conformation thus prevents the hammerhead ribozyme from self-cleavage, ...
... mRNA transcript and preventing expression of the gene. However, when the desired ligand binds to the aptamer region, a conformation change occurs that disrupts the active conformation of the hammerhead ribozyme. This ligand-bound conformation thus prevents the hammerhead ribozyme from self-cleavage, ...
wanted - Copenhagen Plant Science Centre
... DNA that does not code for proteins (non-coding DNA) makes up the vast majority of bases in many genomes yet we understand little about its role. Non-coding regions are actively transcribed by the same complex transcribing genes (RNA polymerase II, Pol II). Transcription of non-coding sequences resu ...
... DNA that does not code for proteins (non-coding DNA) makes up the vast majority of bases in many genomes yet we understand little about its role. Non-coding regions are actively transcribed by the same complex transcribing genes (RNA polymerase II, Pol II). Transcription of non-coding sequences resu ...
Chapter 2 DNA, RNA, Transcription and Translation I. DNA
... Effector (or called inducer) molecules bind to the repressor and release from the operator region on (induction). e.g. the lacZ gene is off w/o -galactoside, when the substrate is added, the enzyme activity appears within 2-3 min IPTG (isopropyl-beta-Dthiogalactopyranoside), a synthetic analogue ...
... Effector (or called inducer) molecules bind to the repressor and release from the operator region on (induction). e.g. the lacZ gene is off w/o -galactoside, when the substrate is added, the enzyme activity appears within 2-3 min IPTG (isopropyl-beta-Dthiogalactopyranoside), a synthetic analogue ...
Lecture 12
... method identified 10 predicted ESE motifs. Representatives of all 10 motifs were found to display enhancer activity in vivo, whereas point mutants of these sequences exhibited sharply reduced activity. • The motifs identified enable prediction of the splicing phenotypes of exonic mutations in human ...
... method identified 10 predicted ESE motifs. Representatives of all 10 motifs were found to display enhancer activity in vivo, whereas point mutants of these sequences exhibited sharply reduced activity. • The motifs identified enable prediction of the splicing phenotypes of exonic mutations in human ...
Introduction to Biology
... throwing the virus into a predesigned protein soup that contained all the polymerases and other enzymatic ingredients necessary for RNA transcription and translation. The synthetic virus was able to successfully replicate itself from this mixture.” ...
... throwing the virus into a predesigned protein soup that contained all the polymerases and other enzymatic ingredients necessary for RNA transcription and translation. The synthetic virus was able to successfully replicate itself from this mixture.” ...
Transcription and Translation
... • All 3 kinds of RNA are made by Transcription: mRNA, rRNA and tRNA • mRNA – carries the code from DNA to Ribosome • rRNA – makes up the Ribosomes (site of protein production) • tRNA – carries the amino acids to the ribosomes to be made into proteins • Most biology classes focus on the production of ...
... • All 3 kinds of RNA are made by Transcription: mRNA, rRNA and tRNA • mRNA – carries the code from DNA to Ribosome • rRNA – makes up the Ribosomes (site of protein production) • tRNA – carries the amino acids to the ribosomes to be made into proteins • Most biology classes focus on the production of ...
ch 18 reading guide
... Feedback inhibition is a recurring mechanism throughout biological systems. In the case of E. coli regulating tryptophan synthesis, is it positive or negative inhibition? Explain your choice. ...
... Feedback inhibition is a recurring mechanism throughout biological systems. In the case of E. coli regulating tryptophan synthesis, is it positive or negative inhibition? Explain your choice. ...
Gene Section DDX43 (DEAD (Asp-Glu-Ala-Asp) box polypeptide 43) Atlas of Genetics and Cytogenetics
... comparable to that of LB23-SAR. Gene HAGE was found to be expressed in 90 out of 383 tumor samples of different histological types, well above the level in normal tissues; about 5% of the positive samples showed a level of expression above 10% of the level of LB23-SAR, and 7% showed a level of expre ...
... comparable to that of LB23-SAR. Gene HAGE was found to be expressed in 90 out of 383 tumor samples of different histological types, well above the level in normal tissues; about 5% of the positive samples showed a level of expression above 10% of the level of LB23-SAR, and 7% showed a level of expre ...
RNA interference
RNA interference (RNAi) is a biological process in which RNA molecules inhibit gene expression, typically by causing the destruction of specific mRNA molecules. Historically, it was known by other names, including co-suppression, post-transcriptional gene silencing (PTGS), and quelling. Only after these apparently unrelated processes were fully understood did it become clear that they all described the RNAi phenomenon. Andrew Fire and Craig C. Mello shared the 2006 Nobel Prize in Physiology or Medicine for their work on RNA interference in the nematode worm Caenorhabditis elegans, which they published in 1998.Two types of small ribonucleic acid (RNA) molecules – microRNA (miRNA) and small interfering RNA (siRNA) – are central to RNA interference. RNAs are the direct products of genes, and these small RNAs can bind to other specific messenger RNA (mRNA) molecules and either increase or decrease their activity, for example by preventing an mRNA from producing a protein. RNA interference has an important role in defending cells against parasitic nucleotide sequences – viruses and transposons. It also influences development.The RNAi pathway is found in many eukaryotes, including animals, and is initiated by the enzyme Dicer, which cleaves long double-stranded RNA (dsRNA) molecules into short double-stranded fragments of ~20 nucleotide siRNAs. Each siRNA is unwound into two single-stranded RNAs (ssRNAs), the passenger strand and the guide strand. The passenger strand is degraded and the guide strand is incorporated into the RNA-induced silencing complex (RISC). The most well-studied outcome is post-transcriptional gene silencing, which occurs when the guide strand pairs with a complementary sequence in a messenger RNA molecule and induces cleavage by Argonaute, the catalytic component of the RISC complex. In some organisms, this process spreads systemically, despite the initially limited molar concentrations of siRNA.RNAi is a valuable research tool, both in cell culture and in living organisms, because synthetic dsRNA introduced into cells can selectively and robustly induce suppression of specific genes of interest. RNAi may be used for large-scale screens that systematically shut down each gene in the cell, which can help to identify the components necessary for a particular cellular process or an event such as cell division. The pathway is also used as a practical tool in biotechnology, medicine and insecticides.























