Project No: 2
... and in six other Vibrio species after a heat shock from 30 to 42C {764}. 4. Enteropathogenicity of V. parahaemolyticus assayed with Caco2 cells We have been working with Dr. P.S.Liu, a cytologist, in establishing a new cell model for the investigation of the cytotoxic and cytotonic virulence factors ...
... and in six other Vibrio species after a heat shock from 30 to 42C {764}. 4. Enteropathogenicity of V. parahaemolyticus assayed with Caco2 cells We have been working with Dr. P.S.Liu, a cytologist, in establishing a new cell model for the investigation of the cytotoxic and cytotonic virulence factors ...
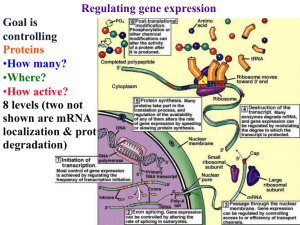
Controlling Gene Expression in Bacteria
... cells all the time. These continually expressed genes are called constitutive genes. Other genes are only needed by certain cells or at specific times. The expression of these inducible genes is tightly controlled. For example, pancreas beta cells make the protein insulin by expressing the insul ...
... cells all the time. These continually expressed genes are called constitutive genes. Other genes are only needed by certain cells or at specific times. The expression of these inducible genes is tightly controlled. For example, pancreas beta cells make the protein insulin by expressing the insul ...
Inquiry into Life Twelfth Edition
... – Prevents formation of a hairpin – This is part of the transcription termination signal which causes attenuation ...
... – Prevents formation of a hairpin – This is part of the transcription termination signal which causes attenuation ...
Foundations of Biology - Geoscience Research Institute
... cells all the time. These continually expressed genes are called constitutive genes. Other genes are only needed by certain cells or at specific times. The expression of these inducible genes is tightly controlled. For example, pancreas beta cells make the protein insulin by expressing the insul ...
... cells all the time. These continually expressed genes are called constitutive genes. Other genes are only needed by certain cells or at specific times. The expression of these inducible genes is tightly controlled. For example, pancreas beta cells make the protein insulin by expressing the insul ...
7.1 The lac Operon
... they have an affinity for each other. Second, CAP and RNA polymerase, when both are bound to their DNA sites, can be chemically cross-linked to each other, suggesting that they are in close proximity. ...
... they have an affinity for each other. Second, CAP and RNA polymerase, when both are bound to their DNA sites, can be chemically cross-linked to each other, suggesting that they are in close proximity. ...
Chapter 24 Translation
... 24.15 Termination Codons Are Recognized by Protein Factors • Termination codons are recognized by protein release factors, not by aminoacyltRNAs. • RF1 – The bacterial release factor that recognizes UAA and UAG as signals to terminate polypeptide translation. • RF2 – The bacterial release factor th ...
... 24.15 Termination Codons Are Recognized by Protein Factors • Termination codons are recognized by protein release factors, not by aminoacyltRNAs. • RF1 – The bacterial release factor that recognizes UAA and UAG as signals to terminate polypeptide translation. • RF2 – The bacterial release factor th ...
Inquiry into Life Twelfth Edition
... – Prevents formation of a hairpin – This is part of the transcription termination signal which causes attenuation ...
... – Prevents formation of a hairpin – This is part of the transcription termination signal which causes attenuation ...
Print
... In mammals, mtDNA is maternally inherited (for references, see Refs. 101, 198). In general, paternal mtDNA is lost during the first few embryonic cell divisions and does not contribute mtDNA to the offspring, although there are reports of the presence of the paternal lineage in somatic tissues (192) ...
... In mammals, mtDNA is maternally inherited (for references, see Refs. 101, 198). In general, paternal mtDNA is lost during the first few embryonic cell divisions and does not contribute mtDNA to the offspring, although there are reports of the presence of the paternal lineage in somatic tissues (192) ...
Transcriptional Paradigms in Mammalian Mitochondrial Biogenesis
... In mammals, mtDNA is maternally inherited (for references, see Refs. 101, 198). In general, paternal mtDNA is lost during the first few embryonic cell divisions and does not contribute mtDNA to the offspring, although there are reports of the presence of the paternal lineage in somatic tissues (192) ...
... In mammals, mtDNA is maternally inherited (for references, see Refs. 101, 198). In general, paternal mtDNA is lost during the first few embryonic cell divisions and does not contribute mtDNA to the offspring, although there are reports of the presence of the paternal lineage in somatic tissues (192) ...
No Slide Title
... 1) CPSF binds AAUAAA in hnRNA 2) CStF binds; CFI, CFII bind in between 3) PAP (PolyA polymerase) binds & cleaves 10-35 b 3’ to ...
... 1) CPSF binds AAUAAA in hnRNA 2) CStF binds; CFI, CFII bind in between 3) PAP (PolyA polymerase) binds & cleaves 10-35 b 3’ to ...
Comparison of Identified TSS Locations to Other
... system process behavior cellular devel. process cell motility ...
... system process behavior cellular devel. process cell motility ...
Molecular Biology of Transcription and RNA Processing
... (see Chapter 7). After a short incubation period to incorporate the labeled nucleotides, a “chase” step replaces any remaining unincorporated radioactive nucleotides by introducing an excess of unlabeled nucleotides. An experimenter can then observe the location and movement of the labeled nucleic a ...
... (see Chapter 7). After a short incubation period to incorporate the labeled nucleotides, a “chase” step replaces any remaining unincorporated radioactive nucleotides by introducing an excess of unlabeled nucleotides. An experimenter can then observe the location and movement of the labeled nucleic a ...
RNA-based regulation of genes of tryptophan synthesis
... Regulatory subtleties abound, however, therefore each event—at each stage—is not absolute. Other regulatory processes influence trp mRNA synthesis and survival, trp coding region translation, trp enzyme function and turnover, and trp enzyme activity (Landick and Yanofsky 1987; Yanofsky and Crawford ...
... Regulatory subtleties abound, however, therefore each event—at each stage—is not absolute. Other regulatory processes influence trp mRNA synthesis and survival, trp coding region translation, trp enzyme function and turnover, and trp enzyme activity (Landick and Yanofsky 1987; Yanofsky and Crawford ...
lac
... doesn’t waste time and energy making mRNA and proteins that are not needed. The lac genes are only transcribed if lactose is present in the growth medium. ...
... doesn’t waste time and energy making mRNA and proteins that are not needed. The lac genes are only transcribed if lactose is present in the growth medium. ...
SB2. Students will analyze how biological traits are passed on to
... protein is called a silent mutation. • Mutation that occurs on the noncoding region (intron) may not affect the encoded protein at all. – Segment of gene that does not code for an amino acid What are some mutagenic factors and what effects do they have on ...
... protein is called a silent mutation. • Mutation that occurs on the noncoding region (intron) may not affect the encoded protein at all. – Segment of gene that does not code for an amino acid What are some mutagenic factors and what effects do they have on ...
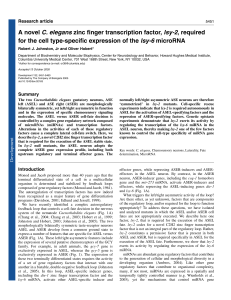
A novel C. elegans zinc finger transcription factor, lsy
... morphologically bilaterally symmetric taste-receptor neurons ASEL and ASER develop from a common ground state to express a number of features that are specific for ASEL versus ASER (Fig. 1A). These left/right asymmetric features include the expression of several putative chemoreceptors of the GCY fa ...
... morphologically bilaterally symmetric taste-receptor neurons ASEL and ASER develop from a common ground state to express a number of features that are specific for ASEL versus ASER (Fig. 1A). These left/right asymmetric features include the expression of several putative chemoreceptors of the GCY fa ...
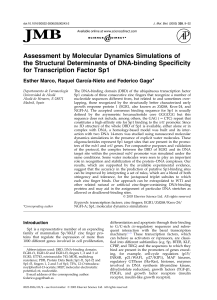
Assessment by Molecular Dynamics Simulations of the Structural
... structures available for Sp1 are those of individual zinc fingers 2 and 3 in solution.41 Sequence alignment of mammalian Sp1, WT1, and other zinc finger-containing transcription factors such as EGR1 reveals a tandem array of three Cys2-His2 zinc finger domains in their C-terminal regions (Figure 1). ...
... structures available for Sp1 are those of individual zinc fingers 2 and 3 in solution.41 Sequence alignment of mammalian Sp1, WT1, and other zinc finger-containing transcription factors such as EGR1 reveals a tandem array of three Cys2-His2 zinc finger domains in their C-terminal regions (Figure 1). ...
Chapter 15
... • Francis Crick and Sydney Brenner determined how the order of nucleotides in DNA encoded amino acid order • Codon – block of 3 DNA nucleotides corresponding to an amino acid • Introduced single nulcleotide insertions or deletions and looked for mutations ...
... • Francis Crick and Sydney Brenner determined how the order of nucleotides in DNA encoded amino acid order • Codon – block of 3 DNA nucleotides corresponding to an amino acid • Introduced single nulcleotide insertions or deletions and looked for mutations ...
Unusual C-terminal domain of the largest subunit of RNA
... considerably from this consensus sequence, but the basic structure observed in other higher eukaryotes is still present (reviewed in (2,10). Three characteristics of the C-terminal domain in eukaryotes suggest that this region might form a unique secondary structure: (i) the amino acid composition, ...
... considerably from this consensus sequence, but the basic structure observed in other higher eukaryotes is still present (reviewed in (2,10). Three characteristics of the C-terminal domain in eukaryotes suggest that this region might form a unique secondary structure: (i) the amino acid composition, ...
Transcription
... • Polymerase II (pol2 for short): transcribes protein-coding genes. We will mainly be discussing this enzyme • Polymerase III: transcribes the 5S ribosomal RNA genes, transfer RNA genes, and small non-coding RNA genes. • Mitochondria and chloroplasts also have their own separate RNA ...
... • Polymerase II (pol2 for short): transcribes protein-coding genes. We will mainly be discussing this enzyme • Polymerase III: transcribes the 5S ribosomal RNA genes, transfer RNA genes, and small non-coding RNA genes. • Mitochondria and chloroplasts also have their own separate RNA ...
GCF (T-14): sc-133419 - Santa Cruz Biotechnology
... GCF (GC-rich sequence DNA-binding factor), also known as C2orf3 (chromosome 2 open reading frame 3), transcription factor 9 (TCF-9) or DNABF, is a 781 amino acid nuclear protein that belongs to the GCF family. Widely expressed, GCF binds the GC-rich sequences of β-Actin, EGFR and calciumdependent pr ...
... GCF (GC-rich sequence DNA-binding factor), also known as C2orf3 (chromosome 2 open reading frame 3), transcription factor 9 (TCF-9) or DNABF, is a 781 amino acid nuclear protein that belongs to the GCF family. Widely expressed, GCF binds the GC-rich sequences of β-Actin, EGFR and calciumdependent pr ...
current micro 40/5 - Bashan Foundation
... RNA preparation. The heterocyst fraction was controlled microscopically and revealed no contaminating vegetative cells. Isolation of total RNA. Total RNA from both A. variabilis and A. nidulans was isolated by a combination of different standard techniques with the RNeasy Kit supplied by Qiagen (D-H ...
... RNA preparation. The heterocyst fraction was controlled microscopically and revealed no contaminating vegetative cells. Isolation of total RNA. Total RNA from both A. variabilis and A. nidulans was isolated by a combination of different standard techniques with the RNeasy Kit supplied by Qiagen (D-H ...
Transcription factor
In molecular biology and genetics, a transcription factor (sometimes called a sequence-specific DNA-binding factor) is a protein that binds to specific DNA sequences, thereby controlling the rate of transcription of genetic information from DNA to messenger RNA. Transcription factors perform this function alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment of RNA polymerase (the enzyme that performs the transcription of genetic information from DNA to RNA) to specific genes.A defining feature of transcription factors is that they contain one or more DNA-binding domains (DBDs), which attach to specific sequences of DNA adjacent to the genes that they regulate. Additional proteins such as coactivators, chromatin remodelers, histone acetylases, deacetylases, kinases, and methylases, while also playing crucial roles in gene regulation, lack DNA-binding domains, and, therefore, are not classified as transcription factors.