The Anaerobic (Class III) Ribonucleotide Reductase from Lactococcus lactis
... mutants in the L. lactis nrdD gene were still able to grow well under standard anaerobic growth conditions and then overproduced the NrdEF proteins (3). There are three classes of ribonucleotide reductases that differ in their protein structure (see recent reviews in Refs. 2 and 5– 8). All operate v ...
... mutants in the L. lactis nrdD gene were still able to grow well under standard anaerobic growth conditions and then overproduced the NrdEF proteins (3). There are three classes of ribonucleotide reductases that differ in their protein structure (see recent reviews in Refs. 2 and 5– 8). All operate v ...
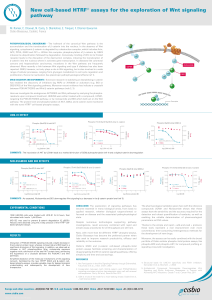
New cell-based HTRF® assays for the exploration of Wnt signaling
... APC, PP2A, GSK3 and CK1 α. Within this complex, phosphorylation of β-catenin by GSK3 induces its ubiquitination followed by degradation. Conversely, binding of Wnt to its frizzled receptor leads to the disruption of the destruction complex, inducing the translocation of β-catenin into the nucleus wh ...
... APC, PP2A, GSK3 and CK1 α. Within this complex, phosphorylation of β-catenin by GSK3 induces its ubiquitination followed by degradation. Conversely, binding of Wnt to its frizzled receptor leads to the disruption of the destruction complex, inducing the translocation of β-catenin into the nucleus wh ...
9/12
... Lophotrichous = tuft at one or both ends Peritrichous = spread evenly over the entire surface Arrangement can be useful in identification ...
... Lophotrichous = tuft at one or both ends Peritrichous = spread evenly over the entire surface Arrangement can be useful in identification ...
A Lipid Transfer–like Protein Is Necessary for Lily
... moving animal cells (Burridge and Chrzanowska-Wodnicka, 1996), are observed (Jauh and Lord, 1995). These configurations were first described in lily by Pierson et al. (1986) and are not detectable in pollen tubes grown in vitro. The adhesion events, the faster growth rates of pollen tubes, and the F ...
... moving animal cells (Burridge and Chrzanowska-Wodnicka, 1996), are observed (Jauh and Lord, 1995). These configurations were first described in lily by Pierson et al. (1986) and are not detectable in pollen tubes grown in vitro. The adhesion events, the faster growth rates of pollen tubes, and the F ...
Facing extremes: archaeal surface-layer (glyco)proteins
... S-layer glycoproteins (Konrad & Eichler, 2002). A combination of pulse–chase radiolabelling and subcellular fractionation approaches has shown that lipid modification of the Haloferax volcanii S-layer glycoprotein only occurs once the S-layer glycoprotein has been delivered across the plasma membran ...
... S-layer glycoproteins (Konrad & Eichler, 2002). A combination of pulse–chase radiolabelling and subcellular fractionation approaches has shown that lipid modification of the Haloferax volcanii S-layer glycoprotein only occurs once the S-layer glycoprotein has been delivered across the plasma membran ...
Recent Advances in Target Characterization and Identification by
... γ-Secretase is an integral membrane protease that cleaves the amyloid precursor proteins (APP) to release Aβ peptides, which have a causative role in the pathogenesis of Alzheimer’s disease (AD) [19,20]. γ-Secretase is a complex of four different integral membrane proteins (presenilin, nicastrin, Ap ...
... γ-Secretase is an integral membrane protease that cleaves the amyloid precursor proteins (APP) to release Aβ peptides, which have a causative role in the pathogenesis of Alzheimer’s disease (AD) [19,20]. γ-Secretase is a complex of four different integral membrane proteins (presenilin, nicastrin, Ap ...
Import of ADP/ATP Carrier into Mitochondria: Two Receptors Act in
... mM MgC12, and 10 mM MOPS/KOH (pH 7.2) (Pfanner and Neupert, 1987; Pfanner et ai., 1987b; S611neret al., 1989, 1990); treatment of mitochondria with trypsin, proteinase K, or elastase (Planner and Neupert, 1987; Pfaller et al., 1988; S611neret al., 1989); SDS-PAGE, transfer of proteins to nitroeellnl ...
... mM MgC12, and 10 mM MOPS/KOH (pH 7.2) (Pfanner and Neupert, 1987; Pfanner et ai., 1987b; S611neret al., 1989, 1990); treatment of mitochondria with trypsin, proteinase K, or elastase (Planner and Neupert, 1987; Pfaller et al., 1988; S611neret al., 1989); SDS-PAGE, transfer of proteins to nitroeellnl ...
Nucleolus-like body of mouse oocytes contains lamin A and B and
... are archetypal nucleic acid remodelling enzymes that, using ATP as a cofactor, can varyingly add or remove DNA supercoils and either form or unlink DNA tangles [30]. Several studies on vertebrate systems indicate that this enzyme plays a role in the shaping of mitotic chromatin: topo II is the main ...
... are archetypal nucleic acid remodelling enzymes that, using ATP as a cofactor, can varyingly add or remove DNA supercoils and either form or unlink DNA tangles [30]. Several studies on vertebrate systems indicate that this enzyme plays a role in the shaping of mitotic chromatin: topo II is the main ...
as a PDF
... Purity analysis of isolated phosphopeptides The purity of each isolated phosphopeptide was analyzed by the method used in two-dimensional peptide mapping as described by Boyle et al . (1991) . In brief, the phosphopeptide peaks resolved from HPLC as described above were collected separately and conc ...
... Purity analysis of isolated phosphopeptides The purity of each isolated phosphopeptide was analyzed by the method used in two-dimensional peptide mapping as described by Boyle et al . (1991) . In brief, the phosphopeptide peaks resolved from HPLC as described above were collected separately and conc ...
Molecular mechanisms of complement evasion: learning from
... cofactor protein (MCP; also known as CD46), complement receptor type 1 (CR1; also known as CD35) and complement receptor of the immunoglobulin superfamily (CRIg; also known as VSIG4), or markers (often polyanionic structures) that attract soluble complement regulators such as factor H (which downreg ...
... cofactor protein (MCP; also known as CD46), complement receptor type 1 (CR1; also known as CD35) and complement receptor of the immunoglobulin superfamily (CRIg; also known as VSIG4), or markers (often polyanionic structures) that attract soluble complement regulators such as factor H (which downreg ...
Protein Structure Prediction Based on Neural Networks
... prediction becomes complex, when the homology-based modelling applies to predicting proteins with similar structures but different chemical makeups, and thus, it requires more computing time [3]. In contrast, amino acid sequences may be acquired through very efficient automated, high throughput expe ...
... prediction becomes complex, when the homology-based modelling applies to predicting proteins with similar structures but different chemical makeups, and thus, it requires more computing time [3]. In contrast, amino acid sequences may be acquired through very efficient automated, high throughput expe ...
Plant lipid transfer proteins - Evolution, expression and function Monika Edstam
... Abstract ............................................................................................................................................................... - 1 Populärvetenskaplig sammanfattning ............................................................................................ ...
... Abstract ............................................................................................................................................................... - 1 Populärvetenskaplig sammanfattning ............................................................................................ ...
How do potentials derived from structural databases relate to true
... not be any evolutionary pressure to make the protein even more stable (Serrano et al., 1993). In summary, problems apparently exist in both of the potential derivation methods. The derived potentials and the true potentials correlate to some extent, but it is not simply a linear relationship, as exp ...
... not be any evolutionary pressure to make the protein even more stable (Serrano et al., 1993). In summary, problems apparently exist in both of the potential derivation methods. The derived potentials and the true potentials correlate to some extent, but it is not simply a linear relationship, as exp ...
Two decades of studying non-covalent biomolecular assemblies by
... With the technology allowing the transmission of intact protein assemblies through the mass spectrometer and their mass measurement with unparalleled accuracy, attention shifted towards devising means for their gasphase disassembly, such that their constituents might be probed. Multiple activation a ...
... With the technology allowing the transmission of intact protein assemblies through the mass spectrometer and their mass measurement with unparalleled accuracy, attention shifted towards devising means for their gasphase disassembly, such that their constituents might be probed. Multiple activation a ...
Field Guide to Protein Folds
... The cyclin-box is an approximately 100-residue domain found in all cyclin and cyclin-like domains that acts as a generalized adaptor motif to recognize diverse proteins and DNAs that are involved in cell cycle and transcriptional regulation. It consists of five helices with a central helix (helix 3) ...
... The cyclin-box is an approximately 100-residue domain found in all cyclin and cyclin-like domains that acts as a generalized adaptor motif to recognize diverse proteins and DNAs that are involved in cell cycle and transcriptional regulation. It consists of five helices with a central helix (helix 3) ...
Systematic Structure-Function Analysis of the Small GTPase Arf1 in Yeast.
... Previous mutagenesis studies have provided valuable information about particular residues such as those involved in nucleotide binding and N-myristoylation and crystallographic studies have identified binding sites for ArfGEF and ArfGAP (Kahn et al. 1995; Goldberg, 1998, 1999; Yahara et al., 2001). ...
... Previous mutagenesis studies have provided valuable information about particular residues such as those involved in nucleotide binding and N-myristoylation and crystallographic studies have identified binding sites for ArfGEF and ArfGAP (Kahn et al. 1995; Goldberg, 1998, 1999; Yahara et al., 2001). ...
prions lecture notes
... - CJD has cerebral involvement so dementia is more common; patient seldom survives a year - GSS is distinct from CJD characterized by cerebellar ataxia and concomitant motor problems, dementia is less common (disease course lasts several years before ultimate death) - fatal insomnias present with an ...
... - CJD has cerebral involvement so dementia is more common; patient seldom survives a year - GSS is distinct from CJD characterized by cerebellar ataxia and concomitant motor problems, dementia is less common (disease course lasts several years before ultimate death) - fatal insomnias present with an ...
Enzymatic Oxygen Scavenging for Photostability without pH Drop in
... (PCD),7,8 with β-D-glucose and 3,4-protocatechuic acid (PCA) as substrate, respectively. However, in both cases, the substrate oxidation produces carboxylic acids (D-gluconic acid and 3,3-carboxy-cis,cismuconic acid, respectively, Figure 1a, reactions 1 and 2). If not buffered sufficiently, this acid p ...
... (PCD),7,8 with β-D-glucose and 3,4-protocatechuic acid (PCA) as substrate, respectively. However, in both cases, the substrate oxidation produces carboxylic acids (D-gluconic acid and 3,3-carboxy-cis,cismuconic acid, respectively, Figure 1a, reactions 1 and 2). If not buffered sufficiently, this acid p ...
F-actin Sequesters Elongation Factor from Interaction with
... min to form ternary complex at room temperature. The assays were performed by using a fluorescence spectrophotometer (model F-2000; Hitachi Sci. Instrs., Mountain View, CA) with 600-nm excitation and emission wavelength at a band pass of 5 nm. Data were collected and analyzed by using the computer s ...
... min to form ternary complex at room temperature. The assays were performed by using a fluorescence spectrophotometer (model F-2000; Hitachi Sci. Instrs., Mountain View, CA) with 600-nm excitation and emission wavelength at a band pass of 5 nm. Data were collected and analyzed by using the computer s ...
Ube2W conjugates ubiquitin to α-amino groups of protein N
... value of 1 000 000). Resolution for HCD spectra was set to 17 500 at m/z 200. Raw MS data files were processed by MaxQuant (version 1.3.0.5) [10,11]. Data were searched against an entire human proteome database plus one containing the known recombinant proteins in the reactions either with or withou ...
... value of 1 000 000). Resolution for HCD spectra was set to 17 500 at m/z 200. Raw MS data files were processed by MaxQuant (version 1.3.0.5) [10,11]. Data were searched against an entire human proteome database plus one containing the known recombinant proteins in the reactions either with or withou ...
RIBOSOME-INACTIVATING PROTEINS: A Plant Perspective
... only by tRNATrp from mammalian and avian cells and not by other tRNAs or tRNATrp from yeast (20–23). Given the variety of assays and assay conditions used for RIP analysis, some differences observed in previous studies may be explained by cofactor requirements. It is not known, however, whether the ...
... only by tRNATrp from mammalian and avian cells and not by other tRNAs or tRNATrp from yeast (20–23). Given the variety of assays and assay conditions used for RIP analysis, some differences observed in previous studies may be explained by cofactor requirements. It is not known, however, whether the ...
Nuclear magnetic resonance spectroscopy of proteins
Nuclear magnetic resonance spectroscopy of proteins (usually abbreviated protein NMR) is a field of structural biology in which NMR spectroscopy is used to obtain information about the structure and dynamics of proteins, and also nucleic acids, and their complexes. The field was pioneered by Richard R. Ernst and Kurt Wüthrich at the ETH, and by Ad Bax, Marius Clore and Angela Gronenborn at the NIH, among others. Structure determination by NMR spectroscopy usually consists of several phases, each using a separate set of highly specialized techniques. The sample is prepared, measurements are made, interpretive approaches are applied, and a structure is calculated and validated.NMR involves the quantum mechanical properties of the central core (""nucleus"") of the atom. These properties depend on the local molecular environment, and their measurement provides a map of how the atoms are linked chemically, how close they are in space, and how rapidly they move with respect to each other. These properties are fundamentally the same as those used in the more familiar Magnetic Resonance Imaging (MRI), but the molecular applications use a somewhat different approach, appropriate to the change of scale from millimeters (of interest to radiologists) to nano-meters (bonded atoms are typically a fraction of a nano-meter apart), a factor of a million. This change of scale requires much higher sensitivity of detection and stability for long term measurement. In contrast to MRI, structural biology studies do not directly generate an image, but rely on complex computer calculations to generate three-dimensional molecular models.Currently most samples are examined in a solution in water, but methods are being developed to also work with solid samples. Data collection relies on placing the sample inside a powerful magnet, sending radio frequency signals through the sample, and measuring the absorption of those signals. Depending on the environment of atoms within the protein, the nuclei of individual atoms will absorb different frequencies of radio signals. Furthermore the absorption signals of different nuclei may be perturbed by adjacent nuclei. This information can be used to determine the distance between nuclei. These distances in turn can be used to determine the overall structure of the protein.A typical study might involve how two proteins interact with each other, possibly with a view to developing small molecules that can be used to probe the normal biology of the interaction (""chemical biology"") or to provide possible leads for pharmaceutical use (drug development). Frequently, the interacting pair of proteins may have been identified by studies of human genetics, indicating the interaction can be disrupted by unfavorable mutations, or they may play a key role in the normal biology of a ""model"" organism like the fruit fly, yeast, the worm C. elegans, or mice. To prepare a sample, methods of molecular biology are typically used to make quantities by bacterial fermentation. This also permits changing the isotopic composition of the molecule, which is desirable because the isotopes behave differently and provide methods for identifying overlapping NMR signals.