Chapter 15 Review Questions
... a protein is its amino acid chain, bonded together with peptide bonds (amide linkages). The secondary structure of a protein begins to shape the amino acid chain using hydrogen bonding, forming alpha-helix and beta-pleated sheet structures. The tertiary structure of a protein gives it 3 dimensions. ...
... a protein is its amino acid chain, bonded together with peptide bonds (amide linkages). The secondary structure of a protein begins to shape the amino acid chain using hydrogen bonding, forming alpha-helix and beta-pleated sheet structures. The tertiary structure of a protein gives it 3 dimensions. ...
Final Exam KEY
... c. (2) Suppose you succeeded in increasing the yield of fragments compared to sample B. Draw in the results on the gel in lane "C". d. (3) On a different day, you realize you forgot to add one of the primers to the tube. You stop the PCR after cycle 13, add the second primer and then let the PCR fin ...
... c. (2) Suppose you succeeded in increasing the yield of fragments compared to sample B. Draw in the results on the gel in lane "C". d. (3) On a different day, you realize you forgot to add one of the primers to the tube. You stop the PCR after cycle 13, add the second primer and then let the PCR fin ...
Phase I: Computational Procedures: I. Measure original band
... A. This was just done at UC Irvine in March of 2013 with excellent results. This process was completed with the aid of Tom Fielder, scientist at UCI, and hasn’t been fully documented yet. It ...
... A. This was just done at UC Irvine in March of 2013 with excellent results. This process was completed with the aid of Tom Fielder, scientist at UCI, and hasn’t been fully documented yet. It ...
Ch. 16 – Control of Gene Expression Sample Questions
... multicellular organisms? A.Grow and divide rapidly B.Cells adjust quickly to outside environment C.Homeostasis D.Quickly synthesize amount and type of enzymes according to available nutrients E.Respond by gene action to oxygen availability ...
... multicellular organisms? A.Grow and divide rapidly B.Cells adjust quickly to outside environment C.Homeostasis D.Quickly synthesize amount and type of enzymes according to available nutrients E.Respond by gene action to oxygen availability ...
Document
... The DNA chips do not have very long shelf life, which proves to be another major disadvantage of the technology. ...
... The DNA chips do not have very long shelf life, which proves to be another major disadvantage of the technology. ...
Nucleotide sequence of the gene encoding the
... C terminus of the component, chromosomal DNA was digested with Bell, circularized by ligation and subjected to an inverse polymerase chain reaction (7) using two oligonucleotides complementary to the 3'-end of the 7.6 kbp PstI fragment. The product, 800 bp in length, contained the sequence encoding ...
... C terminus of the component, chromosomal DNA was digested with Bell, circularized by ligation and subjected to an inverse polymerase chain reaction (7) using two oligonucleotides complementary to the 3'-end of the 7.6 kbp PstI fragment. The product, 800 bp in length, contained the sequence encoding ...
DNA Code problerm
... 3. Which of the following is not true about eukaryotic DNA? A. It is an exceedingly long and fragile molecule. B. It is packaged into successively compact formations. C. The entire molecule has encoded information for protein synthesis. D. In the condensed form, it is transcriptionally inactive. E. ...
... 3. Which of the following is not true about eukaryotic DNA? A. It is an exceedingly long and fragile molecule. B. It is packaged into successively compact formations. C. The entire molecule has encoded information for protein synthesis. D. In the condensed form, it is transcriptionally inactive. E. ...
RNA Polymerase II analysis in Drosophila Melanogaster
... Most of the differences in nucleotides between organisms are situated in noncoding DNA regions. These non coding regions affect the expression levels of genes thus making phenotypes depending more on differential expression rather than genes mutation. This project aim is to study the behaviour of RN ...
... Most of the differences in nucleotides between organisms are situated in noncoding DNA regions. These non coding regions affect the expression levels of genes thus making phenotypes depending more on differential expression rather than genes mutation. This project aim is to study the behaviour of RN ...
Slide ()
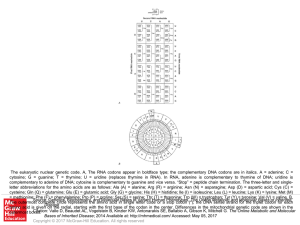
... The eukaryotic nuclear genetic code. A, The RNA codons appear in boldface type; the complementary DNA codons are in italics. A = adenine; C = cytosine; G = guanine; T = thymine; U = uridine (replaces thymine in RNA). In RNA, adenine is complementary to thymine of DNA; uridine is complementary to ade ...
... The eukaryotic nuclear genetic code. A, The RNA codons appear in boldface type; the complementary DNA codons are in italics. A = adenine; C = cytosine; G = guanine; T = thymine; U = uridine (replaces thymine in RNA). In RNA, adenine is complementary to thymine of DNA; uridine is complementary to ade ...
summing-up - Zanichelli online per la scuola
... synthesis of the strands is DNA polymerase, which requires a fuse consisting of a short fragment of RNA (known as a primer). The DNA polymerase is able to synthesise only in the direction 3’-5’ of the template ...
... synthesis of the strands is DNA polymerase, which requires a fuse consisting of a short fragment of RNA (known as a primer). The DNA polymerase is able to synthesise only in the direction 3’-5’ of the template ...
Bartlett`s Lecture
... likely to yield good PCR • Amino acid racemization: conversion of L-amino acids to D-amino acids • Rate depends on water, temperature, chelated metal ions (things that also affect rate of DNA depurination) • The higher the D/L ratio, the less likely that DNA can be isolated: >0.08, no DNA will be is ...
... likely to yield good PCR • Amino acid racemization: conversion of L-amino acids to D-amino acids • Rate depends on water, temperature, chelated metal ions (things that also affect rate of DNA depurination) • The higher the D/L ratio, the less likely that DNA can be isolated: >0.08, no DNA will be is ...
biology 1 - Saddleback College
... • Know how genes are produced (copied) in mass quantities (plasmids of bacteria). • Know the difference between PCR and RFLP • What are restriction enzymes? Would PCR or RFLP utilize restriction enzymes? • Northern, Southern & Western blotting – what are they testing for. • Gene cloning – know how a ...
... • Know how genes are produced (copied) in mass quantities (plasmids of bacteria). • Know the difference between PCR and RFLP • What are restriction enzymes? Would PCR or RFLP utilize restriction enzymes? • Northern, Southern & Western blotting – what are they testing for. • Gene cloning – know how a ...
LAB 2 LECTURE The Molecular Basis for Species Diversity DNA
... b. Guanine (G) always pairs with Cytosine (C) • A gene is a segment of DNA, or a subset of bases within this long sequence of bases. III. Making copies of DNA – Replication 1. The two strands unzip and separate from each other, and are then copied. IV. Proteins 1. There are structural proteins and r ...
... b. Guanine (G) always pairs with Cytosine (C) • A gene is a segment of DNA, or a subset of bases within this long sequence of bases. III. Making copies of DNA – Replication 1. The two strands unzip and separate from each other, and are then copied. IV. Proteins 1. There are structural proteins and r ...
Real-time polymerase chain reaction

A real-time polymerase chain reaction is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). It monitors the amplification of a targeted DNA molecule during the PCR, i.e. in real-time, and not at its end, as in conventional PCR. Real-time PCR can be used quantitatively (Quantitative real-time PCR), semi-quantitatively, i.e. above/below a certain amount of DNA molecules (Semi quantitative real-time PCR) or qualitatively (Qualitative real-time PCR).Two common methods for the detection of PCR products in real-time PCR are: (1) non-specific fluorescent dyes that intercalate with any double-stranded DNA, and (2) sequence-specific DNA probes consisting of oligonucleotides that are labelled with a fluorescent reporter which permits detection only after hybridization of the probe with its complementary sequence.The Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines propose that the abbreviation qPCR be used for quantitative real-time PCR and that RT-qPCR be used for reverse transcription–qPCR [1]. The acronym ""RT-PCR"" commonly denotes reverse transcription polymerase chain reaction and not real-time PCR, but not all authors adhere to this convention.