Macromolecules of life: Structure-function and Bioinformatics 356
... recognition and interactions. Bioinformatics predictions of protein and small molecule DNA interactions. Chemical reactivity of amino acids. Domain structures of proteins and Ramachandran plots. Protein folding, sequence motifs and domains, higher order and supramolecular structure, self-assembly, c ...
... recognition and interactions. Bioinformatics predictions of protein and small molecule DNA interactions. Chemical reactivity of amino acids. Domain structures of proteins and Ramachandran plots. Protein folding, sequence motifs and domains, higher order and supramolecular structure, self-assembly, c ...
Protein Needs for Athletes
... your diet to provide all the EAAs. • Milk is effective at stimulating muscle growth following training exercise. Consider drinking a glass of low-fat chocolate milk after a workout. ...
... your diet to provide all the EAAs. • Milk is effective at stimulating muscle growth following training exercise. Consider drinking a glass of low-fat chocolate milk after a workout. ...
Amino acids
... globular proteins assume as a consequence of the noncovalent interactions between the side chains in their primary structure) • quarternary (Many proteins are composed of several polypeptide chains = subunits) ...
... globular proteins assume as a consequence of the noncovalent interactions between the side chains in their primary structure) • quarternary (Many proteins are composed of several polypeptide chains = subunits) ...
Knuffke Prezi- Macromolecules
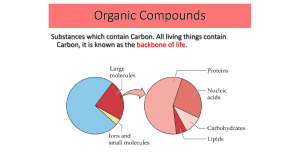
... Substances which contain Carbon. All living things contain Carbon, it is known as the backbone of life. ...
... Substances which contain Carbon. All living things contain Carbon, it is known as the backbone of life. ...
R032 Publication Only Basic Science: Biofilm Key proteins of
... virulent. The proteomic analysis was performed by two-dimensional polyacrylamide gel electrophoresis including sample preparation, isoelectric focusing, electrophoretic separation and image scanning and analysis. The gels were analyzed by software ImageMaster 2D Plattinum, allowing to estimate the i ...
... virulent. The proteomic analysis was performed by two-dimensional polyacrylamide gel electrophoresis including sample preparation, isoelectric focusing, electrophoretic separation and image scanning and analysis. The gels were analyzed by software ImageMaster 2D Plattinum, allowing to estimate the i ...
Mass spectrometric characterization of a conformational epitope of
... bioaffinity- MS show broad bioanalytical potential for direct interaction studies from biological material, as diverse as antigen-antibody and lectin- carbohydrate complexes; affinity binding constants (KD) are determined from milli- to nanomolar ranges [2, 3]. Recent applications of the online-bios ...
... bioaffinity- MS show broad bioanalytical potential for direct interaction studies from biological material, as diverse as antigen-antibody and lectin- carbohydrate complexes; affinity binding constants (KD) are determined from milli- to nanomolar ranges [2, 3]. Recent applications of the online-bios ...
Abstract
... Inferring direct couplings to unveil coevolutionary signals in protein 3D structure, interactions and recognition in signaling networks. Modern sequencing technologies provide us with a rich source of data about the evolutionary history of proteins. Inferring a joint probability distribution of amin ...
... Inferring direct couplings to unveil coevolutionary signals in protein 3D structure, interactions and recognition in signaling networks. Modern sequencing technologies provide us with a rich source of data about the evolutionary history of proteins. Inferring a joint probability distribution of amin ...
Heller’s-ring-test
... aromatic amino acids. It can be obtained by treating the proteins with other acids like HCl and H2SO4. ...
... aromatic amino acids. It can be obtained by treating the proteins with other acids like HCl and H2SO4. ...
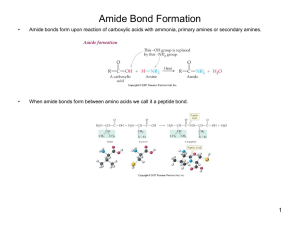
Amide Bond Formation
... Amide bonds form upon reaction of carboxylic acids with ammonia, primary amines or secondary amines. ...
... Amide bonds form upon reaction of carboxylic acids with ammonia, primary amines or secondary amines. ...
Kay Hofmann - Tresch Group
... for every possible peptide. Possible peptides are taken from a proteome-wide sequence database, taking the cleaving enyzme into account. Even if some ions are missing or too much, the correcpt peptide can be identified by a good correlation of expected and observed pattern. Problems with polymorphis ...
... for every possible peptide. Possible peptides are taken from a proteome-wide sequence database, taking the cleaving enyzme into account. Even if some ions are missing or too much, the correcpt peptide can be identified by a good correlation of expected and observed pattern. Problems with polymorphis ...
y-ion series=A, AA, LAA, SLAA
... MS-based Quantitative Methods • Precursor: Quantitation based on the relative intensities of extracted ion chromatograms (XICs) for precursors within a single data set. This is a widely used approach, which can be used with any chemistry that creates a precursor mass shift. For example, 18O, AQUA, ...
... MS-based Quantitative Methods • Precursor: Quantitation based on the relative intensities of extracted ion chromatograms (XICs) for precursors within a single data set. This is a widely used approach, which can be used with any chemistry that creates a precursor mass shift. For example, 18O, AQUA, ...
A new strategy for quantitative proteomics using isotope
... methodology have shown to be powerful alternatives to comparative 2D gel imaging analysis. Nevertheless, these methods also have their limitations. Here we describe a new method termed Isotope Coded Protein Label (ICPL) which is based on isotopic labelling of all free amino groups in proteins. Compa ...
... methodology have shown to be powerful alternatives to comparative 2D gel imaging analysis. Nevertheless, these methods also have their limitations. Here we describe a new method termed Isotope Coded Protein Label (ICPL) which is based on isotopic labelling of all free amino groups in proteins. Compa ...
analysis of a local huntington protein interaction network
... protein. This study sheds light on possible functions for the huntingtin protein though analysis of a local protein-protein interaction network consisting of the huntingtin protein, proteins called primaries that have been found to interact with the huntingtin protein and secondary proteins that int ...
... protein. This study sheds light on possible functions for the huntingtin protein though analysis of a local protein-protein interaction network consisting of the huntingtin protein, proteins called primaries that have been found to interact with the huntingtin protein and secondary proteins that int ...
distinct format
... proteins of which 714 proteins were identified in asexual blood stages (left panel), 931 in gametocytes (right panel) and 645 in gametes. The last two groups provide insights into the biology of the sexual stages of the parasite, and include conserved, stage-specific, secreted and membrane-associate ...
... proteins of which 714 proteins were identified in asexual blood stages (left panel), 931 in gametocytes (right panel) and 645 in gametes. The last two groups provide insights into the biology of the sexual stages of the parasite, and include conserved, stage-specific, secreted and membrane-associate ...
Proteomics - University of Warwick
... At Warwick staff in the Proteomics Research Technology Platform are experts in mass spectrometry based proteomics which investigate the proteomes/proteins and their functions. We are here to help you with proteomic experiments. Mass spectrometry Bioinformatics Test Hypothesis* Application Observatio ...
... At Warwick staff in the Proteomics Research Technology Platform are experts in mass spectrometry based proteomics which investigate the proteomes/proteins and their functions. We are here to help you with proteomic experiments. Mass spectrometry Bioinformatics Test Hypothesis* Application Observatio ...
Proteome - Nematode bioinformatics. Analysis tools and data
... • Post-transcriptional and post translational regulation important! ...
... • Post-transcriptional and post translational regulation important! ...
Supplementary Table 1: A complete list of proteins identified with
... Supplementary Table 1: A complete list of proteins identified with two or more peptides using MaxQuant (version 1.2.2.5) from experiments using anti-acetyl-lysine immunoprecipitation and SILAC (stable isotope labeling with amino acids in cell culture) analysis of MOLM-13 cells treated with nutlin-3 ...
... Supplementary Table 1: A complete list of proteins identified with two or more peptides using MaxQuant (version 1.2.2.5) from experiments using anti-acetyl-lysine immunoprecipitation and SILAC (stable isotope labeling with amino acids in cell culture) analysis of MOLM-13 cells treated with nutlin-3 ...
Project description
... mechanism of the protein biosynthesis process. One of the most unclear stages of protein synthesis in cells is translation termination. According to the newest results, many different proteins are the participants of the termination stage. For instance, besides two “canonical” and the well-known ter ...
... mechanism of the protein biosynthesis process. One of the most unclear stages of protein synthesis in cells is translation termination. According to the newest results, many different proteins are the participants of the termination stage. For instance, besides two “canonical” and the well-known ter ...
Title: A Human Tumor Genome Project: From
... normally required to detect secreted proteins from plasma in order to discover the useful cancer biomarkers. Plasma has a wide range of protein complexity, very large number of proteins from most protein components in the body with extensive posttranslational modifications and physiological and temp ...
... normally required to detect secreted proteins from plasma in order to discover the useful cancer biomarkers. Plasma has a wide range of protein complexity, very large number of proteins from most protein components in the body with extensive posttranslational modifications and physiological and temp ...
Construct name
... Data S2 Identification of phosphorylation sites by Mass Spectrometry. The MS analysis was performed in Mass Spectrometry Laboratory, Institute of Biochemistry and Biophysics, Warsaw, Poland. Proteins were IP with HA antibody from protein extract that was isolated form agroinfiltrated N. benthamiana ...
... Data S2 Identification of phosphorylation sites by Mass Spectrometry. The MS analysis was performed in Mass Spectrometry Laboratory, Institute of Biochemistry and Biophysics, Warsaw, Poland. Proteins were IP with HA antibody from protein extract that was isolated form agroinfiltrated N. benthamiana ...
Ch. 5. Protein Purification and Characterization Techniques
... Isoelectric Focusing • Isolectric focusing- based on differing isoelectric pts. (pI) of proteins • Gel is prepared with pH gradient that parallels electricfield. What does this do? • Charge on the protein changes as it migrates. • When it gets to pI, has no charge and stops ...
... Isoelectric Focusing • Isolectric focusing- based on differing isoelectric pts. (pI) of proteins • Gel is prepared with pH gradient that parallels electricfield. What does this do? • Charge on the protein changes as it migrates. • When it gets to pI, has no charge and stops ...
Supporting Information File SF5
... guanidine thiocyanate and phenol in mono-phase solution, the thymus tissues were homogenized in TRI REAGENT (1 ml per 50-100 mg of tissue) in a Polytron homogenizer and light vertex and incubation (10 min, room temp). 0.2 ml of chloroform was added per ml of TRI REAGENT and incubation (10 min, room ...
... guanidine thiocyanate and phenol in mono-phase solution, the thymus tissues were homogenized in TRI REAGENT (1 ml per 50-100 mg of tissue) in a Polytron homogenizer and light vertex and incubation (10 min, room temp). 0.2 ml of chloroform was added per ml of TRI REAGENT and incubation (10 min, room ...
Protein mass spectrometry

Protein mass spectrometry refers to the application of mass spectrometry to the study of proteins. Mass spectrometry is an important emerging method for the characterization of proteins. The two primary methods for ionization of whole proteins are electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI). In keeping with the performance and mass range of available mass spectrometers, two approaches are used for characterizing proteins. In the first, intact proteins are ionized by either of the two techniques described above, and then introduced to a mass analyzer. This approach is referred to as ""top-down"" strategy of protein analysis. In the second, proteins are enzymatically digested into smaller peptides using a protease such as trypsin. Subsequently these peptides are introduced into the mass spectrometer and identified by peptide mass fingerprinting or tandem mass spectrometry. Hence, this latter approach (also called ""bottom-up"" proteomics) uses identification at the peptide level to infer the existence of proteins.Whole protein mass analysis is primarily conducted using either time-of-flight (TOF) MS, or Fourier transform ion cyclotron resonance (FT-ICR). These two types of instrument are preferable here because of their wide mass range, and in the case of FT-ICR, its high mass accuracy. Mass analysis of proteolytic peptides is a much more popular method of protein characterization, as cheaper instrument designs can be used for characterization. Additionally, sample preparation is easier once whole proteins have been digested into smaller peptide fragments. The most widely used instrument for peptide mass analysis are the MALDI time-of-flight instruments as they permit the acquisition of peptide mass fingerprints (PMFs) at high pace (1 PMF can be analyzed in approx. 10 sec). Multiple stage quadrupole-time-of-flight and the quadrupole ion trap also find use in this application.