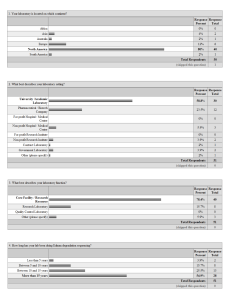
Survey
... c) Good thing considering obsolescence. Parts becoming short supply. d) Can make many repairs in-house - time permitting 31. Which, if any, of the following mass spectrometry based protein analysis techniques does your lab perform; check all that apply a) Nano-ESI de novo sequencing b) LC/M 35. What ...
... c) Good thing considering obsolescence. Parts becoming short supply. d) Can make many repairs in-house - time permitting 31. Which, if any, of the following mass spectrometry based protein analysis techniques does your lab perform; check all that apply a) Nano-ESI de novo sequencing b) LC/M 35. What ...
Polypeptide: alpha-helix and beta
... Description: Models are used to illustrate secondary protein structure. Concept: Peptide chains tend to form orderly hydrogen-bonded arrangements. Materials: alpha-helix and beta-sheet models made by Prof. Ewing Procedure: Models may be used to help explain secondary protein structure. Related Inf ...
... Description: Models are used to illustrate secondary protein structure. Concept: Peptide chains tend to form orderly hydrogen-bonded arrangements. Materials: alpha-helix and beta-sheet models made by Prof. Ewing Procedure: Models may be used to help explain secondary protein structure. Related Inf ...
Protein Purification and Characterization Techniques
... • Salt stabilizes the various charged groups on a protein molecule and enhance the polarity of water, thus attracting protein into the solution and enhancing the solubility of protein • Ammonium sulfate is the most common reagent to use at this step • This technique is important but results are crud ...
... • Salt stabilizes the various charged groups on a protein molecule and enhance the polarity of water, thus attracting protein into the solution and enhancing the solubility of protein • Ammonium sulfate is the most common reagent to use at this step • This technique is important but results are crud ...
Proteomics
... Protein-Protein Interactions • High-throughput strategy – Prediction from sequence • In silico analysis ...
... Protein-Protein Interactions • High-throughput strategy – Prediction from sequence • In silico analysis ...
The Human Proteome
... to an electro-magnetic field whereby it separates them by their mass-to-charge ratio These separated compounds are then measured by a detector Tandem mass spectrometry involves several steps of mass spectrometry with a fragmentation step in between This can be used to fragment proteins over mu ...
... to an electro-magnetic field whereby it separates them by their mass-to-charge ratio These separated compounds are then measured by a detector Tandem mass spectrometry involves several steps of mass spectrometry with a fragmentation step in between This can be used to fragment proteins over mu ...
Biochemistry- Ch 11. Carbohydrates
... cleaves the glycosidic bonds to the sialic acid residues, freeing the virus to infect the cell. Inhibitor of this enzyme are showing some promise as anti-influenza agents. ...
... cleaves the glycosidic bonds to the sialic acid residues, freeing the virus to infect the cell. Inhibitor of this enzyme are showing some promise as anti-influenza agents. ...
Presentation Title Goes Right Here
... ...but protein sequence databases do! Searching traditional protein sequence databases biases the results towards well-understood protein isoforms! ...
... ...but protein sequence databases do! Searching traditional protein sequence databases biases the results towards well-understood protein isoforms! ...
The Nobel Prize in Chemistry 1948 Arne Tiselius
... TESS The Nobel Prize in Chemistry 1948 Arne Tiselius ...
... TESS The Nobel Prize in Chemistry 1948 Arne Tiselius ...
An Introduction to Proteomics
... Along these lines, as with the HGP, when it comes to literature, what do you do, just publish the whole thing? • This is another stumbling block of what to do with all of this information. Proteomics needs its “own PCR,” or “miracle” tool, to increase the throughput. • A new technology, or instr ...
... Along these lines, as with the HGP, when it comes to literature, what do you do, just publish the whole thing? • This is another stumbling block of what to do with all of this information. Proteomics needs its “own PCR,” or “miracle” tool, to increase the throughput. • A new technology, or instr ...
Supplementary Information (doc 50K)
... spray voltage of 2 kV and an automated data dependent MS-MS analysis performed on the top 5 most abundant ions from each MS scan before another full MS scan was performed. Peptides were analyzed once then excluded for 90 seconds. A Mascot generic data file was generated from the MS-MS data using RAW ...
... spray voltage of 2 kV and an automated data dependent MS-MS analysis performed on the top 5 most abundant ions from each MS scan before another full MS scan was performed. Peptides were analyzed once then excluded for 90 seconds. A Mascot generic data file was generated from the MS-MS data using RAW ...
Protein Function Foldable Activity
... which provides the protective structures of our hair and nails. ...
... which provides the protective structures of our hair and nails. ...
Introduction of SILAC and its applications
... Rafts bind not just tyrosine kinases but also serine/threonine kinases and G proteins (more ubiquitous than thought) ...
... Rafts bind not just tyrosine kinases but also serine/threonine kinases and G proteins (more ubiquitous than thought) ...
Key concepts_Protein processing and modification
... complexes called translocons. A number of different mechanisms are employed in bacteria and eukaryotes. In particular, proteins can be translocated either directly from the ribosome, in cotranslational translocation, or from the cytoplasm, in post-translational translocation. The vast majority of pr ...
... complexes called translocons. A number of different mechanisms are employed in bacteria and eukaryotes. In particular, proteins can be translocated either directly from the ribosome, in cotranslational translocation, or from the cytoplasm, in post-translational translocation. The vast majority of pr ...
1 - From protein structure to biological function through interactomics
... (affinity purification followed by quantitative mass spectrometry), new cloning methods such as CPEC and SLiCE, multi-tiered protein expression approaches, in cell NMR, protein binding-sites through cross-linking approaches, ITC, and several other auxiliary biophysical techniques. The course started ...
... (affinity purification followed by quantitative mass spectrometry), new cloning methods such as CPEC and SLiCE, multi-tiered protein expression approaches, in cell NMR, protein binding-sites through cross-linking approaches, ITC, and several other auxiliary biophysical techniques. The course started ...
A dead-end street of protein folding
... Amino acid sequences of globular proteins encode their 3D-structures linked to their biological function. More evidence supports that for many proteins a second, well organized, but quite different 3Dstructure also exists. The latter types of conformers have an architecture similar to the aggregated ...
... Amino acid sequences of globular proteins encode their 3D-structures linked to their biological function. More evidence supports that for many proteins a second, well organized, but quite different 3Dstructure also exists. The latter types of conformers have an architecture similar to the aggregated ...
Protein Electrophoresis
... •Transfer (blot) proteins onto membrane (nylon , nitrocellulose) •Probe the membrane with 1o antibody (recognizes your protein) •Add 2o antibody (this antibody is linked to an enzyme) •Substrate is added ...
... •Transfer (blot) proteins onto membrane (nylon , nitrocellulose) •Probe the membrane with 1o antibody (recognizes your protein) •Add 2o antibody (this antibody is linked to an enzyme) •Substrate is added ...
method for selective purification of sialic acid containing
... Martin Røssel Larsen, Assistent Professor - Department of Biochemistry and Molecular Biology, University of Southern Denmark Martin Røssel Larsen have worked in the area of mass spectrometry and proteomics for the last 12 years, in which he has completed his M.Sc., and Ph.D. degree under the supervi ...
... Martin Røssel Larsen, Assistent Professor - Department of Biochemistry and Molecular Biology, University of Southern Denmark Martin Røssel Larsen have worked in the area of mass spectrometry and proteomics for the last 12 years, in which he has completed his M.Sc., and Ph.D. degree under the supervi ...
TEXT S1- SUPPLEMENTAL METHODS In-solution digestion
... [7]. The cytosolic and membrane protein fractions of both strains were processed independently. Semi-quantitative information was extracted from the LC-MS data and the Mascot search results by extracted ion currents. Details of the IDEAL-Q method are described by Tsou and coworkers [7]. Briefly, IDE ...
... [7]. The cytosolic and membrane protein fractions of both strains were processed independently. Semi-quantitative information was extracted from the LC-MS data and the Mascot search results by extracted ion currents. Details of the IDEAL-Q method are described by Tsou and coworkers [7]. Briefly, IDE ...
Unit1-KA4-Revision
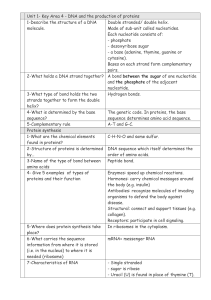
... 2-What holds a DNA strand together? A bond between the sugar of one nucleotide and the phosphate of the adjacent nucleotide. 3-What type of bond holds the two Hydrogen bonds. strands together to form the double helix? 4-What is determined by the base The genetic code. In proteins, the base sequence? ...
... 2-What holds a DNA strand together? A bond between the sugar of one nucleotide and the phosphate of the adjacent nucleotide. 3-What type of bond holds the two Hydrogen bonds. strands together to form the double helix? 4-What is determined by the base The genetic code. In proteins, the base sequence? ...
Proteomics at the Broad Institute Caitlin Feeney, Chemistry and Chemical Biology Introduction Activities
... Cells that we receive are either taken from a specific disease state or treated with variable drugs. Histone proteins are extracted from these cell pellets, digested with trypsin, and the resulting peptides are analyzed by liquid chromatography-mass spectrometric techniques. ...
... Cells that we receive are either taken from a specific disease state or treated with variable drugs. Histone proteins are extracted from these cell pellets, digested with trypsin, and the resulting peptides are analyzed by liquid chromatography-mass spectrometric techniques. ...
105 Quantitative Analysis of Crude Protein
... Quantitative Analysis of Crude Protein (Issued in June 1999) (Updated in November 2013) ...
... Quantitative Analysis of Crude Protein (Issued in June 1999) (Updated in November 2013) ...
Supplementary Methods Quantitative mass spectrometry
... Briefly, peptides (4 µl) were separated by reverse phase chromatography-nLC (Acclaim PepMap RSLC, 1.8 μm x 75 μm x 50 cm) at a flow rate of 250 nL/min over a 90 min ACN gradient from 4 – 40% mobile phase B (A, 0.1% FA; B, 97% ACN in 0.1% FA) and ionized by ESI directly into the mass spectrometer (L ...
... Briefly, peptides (4 µl) were separated by reverse phase chromatography-nLC (Acclaim PepMap RSLC, 1.8 μm x 75 μm x 50 cm) at a flow rate of 250 nL/min over a 90 min ACN gradient from 4 – 40% mobile phase B (A, 0.1% FA; B, 97% ACN in 0.1% FA) and ionized by ESI directly into the mass spectrometer (L ...
Protein mass spectrometry

Protein mass spectrometry refers to the application of mass spectrometry to the study of proteins. Mass spectrometry is an important emerging method for the characterization of proteins. The two primary methods for ionization of whole proteins are electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI). In keeping with the performance and mass range of available mass spectrometers, two approaches are used for characterizing proteins. In the first, intact proteins are ionized by either of the two techniques described above, and then introduced to a mass analyzer. This approach is referred to as ""top-down"" strategy of protein analysis. In the second, proteins are enzymatically digested into smaller peptides using a protease such as trypsin. Subsequently these peptides are introduced into the mass spectrometer and identified by peptide mass fingerprinting or tandem mass spectrometry. Hence, this latter approach (also called ""bottom-up"" proteomics) uses identification at the peptide level to infer the existence of proteins.Whole protein mass analysis is primarily conducted using either time-of-flight (TOF) MS, or Fourier transform ion cyclotron resonance (FT-ICR). These two types of instrument are preferable here because of their wide mass range, and in the case of FT-ICR, its high mass accuracy. Mass analysis of proteolytic peptides is a much more popular method of protein characterization, as cheaper instrument designs can be used for characterization. Additionally, sample preparation is easier once whole proteins have been digested into smaller peptide fragments. The most widely used instrument for peptide mass analysis are the MALDI time-of-flight instruments as they permit the acquisition of peptide mass fingerprints (PMFs) at high pace (1 PMF can be analyzed in approx. 10 sec). Multiple stage quadrupole-time-of-flight and the quadrupole ion trap also find use in this application.