Rhesus ALK-7 / ALK7 / ACVR1C Protein (Fc Tag)
... < 1.0 EU per μg of the protein as determined by the LAL method ...
... < 1.0 EU per μg of the protein as determined by the LAL method ...
Proteins - Kaikoura High School
... • Are polymers • (note: often confusion over peptides, proteins and polypeptides = all are amino acid chains, difference is length.) • Units are amino acids (made up from 64 nucleotide combinations) ...
... • Are polymers • (note: often confusion over peptides, proteins and polypeptides = all are amino acid chains, difference is length.) • Units are amino acids (made up from 64 nucleotide combinations) ...
THE PROTEOME RESPONSE OF LARVAL STAGES OF
... Oysters are one of the most important commercially exploited species cultured in the molluskan hatcheries around the world. Due to rising CO 2 and subsequent decrease in seawater pH, their survival and shell forming processes are threatened globally. Our large-scale CO 2 perturbation experiments at ...
... Oysters are one of the most important commercially exploited species cultured in the molluskan hatcheries around the world. Due to rising CO 2 and subsequent decrease in seawater pH, their survival and shell forming processes are threatened globally. Our large-scale CO 2 perturbation experiments at ...
D6- Bulletin Board Powerful Protein
... • There are 9 essential amino acids that our bodies can’t make, so we need to get them from our food. • If a protein food has all 9 essential amino acids, it is called a complete protein. If it doesn’t, it is called an incomplete protein. • You can eat incomplete protein foods together to make sure ...
... • There are 9 essential amino acids that our bodies can’t make, so we need to get them from our food. • If a protein food has all 9 essential amino acids, it is called a complete protein. If it doesn’t, it is called an incomplete protein. • You can eat incomplete protein foods together to make sure ...
Center for Structural Biology
... solution based on selection by a stationary phase 1. Size- sieve effect, small molecules faster 2. Ion exchange- charge attraction at protein surface Choose “+” stationary phase for proteins with more “-” charge First bind everything, then elute with salt 3. Hydrophobic interaction- hydrophobic ...
... solution based on selection by a stationary phase 1. Size- sieve effect, small molecules faster 2. Ion exchange- charge attraction at protein surface Choose “+” stationary phase for proteins with more “-” charge First bind everything, then elute with salt 3. Hydrophobic interaction- hydrophobic ...
A.P.day52 proteins
... Disulfide linkage: S-H helps hold proteins together to form a tertiary structure. The phosphate group is what makes DNA a negative charged molecule which then can be separated with gel electrophoresis Hydrophobic Van der wall interactions – Nonpolar amino acids avoid water and move to one side and a ...
... Disulfide linkage: S-H helps hold proteins together to form a tertiary structure. The phosphate group is what makes DNA a negative charged molecule which then can be separated with gel electrophoresis Hydrophobic Van der wall interactions – Nonpolar amino acids avoid water and move to one side and a ...
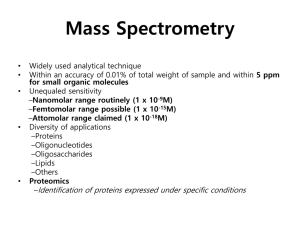
Master course KEMM03 Principles of Mass Spectrometric Protein
... Master course KEMM03 Principles of Mass Spectrometric Protein Characterization ...
... Master course KEMM03 Principles of Mass Spectrometric Protein Characterization ...
CH 908: Mass Spectrometry Lecture 9 Electron Capture Dissociation
... Hofstadler, S. A.; Sannes-Lowery, K. A.; Crooke, S. T.; Ecker, D. J.; Sasmor, H.; Manalili, S.; Griffey, R. H. Multiplexed screening of neutral masstagged RNA targets against ligand libraries with electrospray ionization FTICR MS: A paradigm for high-throughout affinity screening Anal. Chem. ...
... Hofstadler, S. A.; Sannes-Lowery, K. A.; Crooke, S. T.; Ecker, D. J.; Sasmor, H.; Manalili, S.; Griffey, R. H. Multiplexed screening of neutral masstagged RNA targets against ligand libraries with electrospray ionization FTICR MS: A paradigm for high-throughout affinity screening Anal. Chem. ...
A sample for a final examination
... 1. An experimentalist would like to design a simple sequence of alanine and arginine only that will fold into the known structure of lysozyme. He asks his friend (a computational biologist) to estimate the significance of his design (before he is going to do all the hard synthesis work). The compute ...
... 1. An experimentalist would like to design a simple sequence of alanine and arginine only that will fold into the known structure of lysozyme. He asks his friend (a computational biologist) to estimate the significance of his design (before he is going to do all the hard synthesis work). The compute ...
2-D gels and protein mass spectrometry
... c) In protein mass spectrometry, however, the mass spectrum usually consists of one broad peak, centred at a mass corresponding to the atomic weight of the protein, as calculated from the periodic table's atomic masses. For example, consider a hypothetical protein with composition: C601H723N126O230. ...
... c) In protein mass spectrometry, however, the mass spectrum usually consists of one broad peak, centred at a mass corresponding to the atomic weight of the protein, as calculated from the periodic table's atomic masses. For example, consider a hypothetical protein with composition: C601H723N126O230. ...
Protein Engineering
... •Increase the efficiency of enzyme-catalyzed reactions • Eliminate the need for cofactor in enzymatic reaction •Change substrate binding site to increase specificity •Change the thermal tolerance •Change the pH stability •Increase proteins resistance to proteases (purification) •Signal sequences - s ...
... •Increase the efficiency of enzyme-catalyzed reactions • Eliminate the need for cofactor in enzymatic reaction •Change substrate binding site to increase specificity •Change the thermal tolerance •Change the pH stability •Increase proteins resistance to proteases (purification) •Signal sequences - s ...
Elise Young: Animal & Range Sciences
... Linking common factors in the phenomenon of protein clumping observed in several diseases Proteins perform many important functions at the cellular level. However, if proteins do not fold properly, they are prone to aggregating and sticking together, preventing them from performing their functions, ...
... Linking common factors in the phenomenon of protein clumping observed in several diseases Proteins perform many important functions at the cellular level. However, if proteins do not fold properly, they are prone to aggregating and sticking together, preventing them from performing their functions, ...
1. Protein Interactions
... Size: Larger molecules have more active sites Structure: the stability (strength of intramolecular bonds) and molecule unfolding rate ...
... Size: Larger molecules have more active sites Structure: the stability (strength of intramolecular bonds) and molecule unfolding rate ...
Chapter 5 – Proteins and Amino Acids
... 2. Amino Acid Composition 3. High-Quality Proteins 4. Complementary Proteins B. Protein Sparing Nutrition in Practice – Vegetarian Diets A. Are vegetarian diets nutritionally sound? B. What should be my main concerns when planning a nutritionally sound vegetarian diet? C. Isn’t protein a problem in ...
... 2. Amino Acid Composition 3. High-Quality Proteins 4. Complementary Proteins B. Protein Sparing Nutrition in Practice – Vegetarian Diets A. Are vegetarian diets nutritionally sound? B. What should be my main concerns when planning a nutritionally sound vegetarian diet? C. Isn’t protein a problem in ...
Michal Sharon received her BSc in Chemistry from the Hebrew
... Ph.D. studies at the Weizmann Institute of Science focused on studying the three-dimensional structure of proteins by NMR. She conducted postdoctoral research in the Department of Chemistry at the University of Cambridge, UK in the lab of Prof. Carol Robinson until 2007, when she joined the faculty ...
... Ph.D. studies at the Weizmann Institute of Science focused on studying the three-dimensional structure of proteins by NMR. She conducted postdoctoral research in the Department of Chemistry at the University of Cambridge, UK in the lab of Prof. Carol Robinson until 2007, when she joined the faculty ...
011S Product Info
... respectively. Reaction mixtures are quenched with ammonium bicarbonate, combined and analyzed by mass spectrometry. Interpeptide crosslinks will manifest in mass spectra as doublets of signals separated by 8.05 Da, as they contain two N-termini, and non-crosslinked peptides will manifest as doublets ...
... respectively. Reaction mixtures are quenched with ammonium bicarbonate, combined and analyzed by mass spectrometry. Interpeptide crosslinks will manifest in mass spectra as doublets of signals separated by 8.05 Da, as they contain two N-termini, and non-crosslinked peptides will manifest as doublets ...
The_Structure_of_Protein_Activity
... a) The solubility of many proteins in water. b) The precise 3D structure of those enzymes which are proteins. c) The eleasticity of natural fibres such as wool and silk? ...
... a) The solubility of many proteins in water. b) The precise 3D structure of those enzymes which are proteins. c) The eleasticity of natural fibres such as wool and silk? ...
Abstract
... Insoluble β-amyloid peptide (Aβ) deposits formed in the synaptic milieu, chronic activation of glial cells and inflammation are consistent features in Alzheimer’s disease (AD) and strong candidates for the initiation of this process. S100B is one of the numerous pro-inflammatory molecules produced b ...
... Insoluble β-amyloid peptide (Aβ) deposits formed in the synaptic milieu, chronic activation of glial cells and inflammation are consistent features in Alzheimer’s disease (AD) and strong candidates for the initiation of this process. S100B is one of the numerous pro-inflammatory molecules produced b ...
The samples were dissolved in 25 µL of 100 mM ammonium
... Each sample was split equally (10 µL/tube) into 2 tubes. One sample was digested with trypsin and the other tube was digested with LysC. The digestion protocol was the same for both enzymes and involved first diluting the sample 1:1 with 2,2,2-trifluoroethanol, then reducing with DTT at 90°C for 20 ...
... Each sample was split equally (10 µL/tube) into 2 tubes. One sample was digested with trypsin and the other tube was digested with LysC. The digestion protocol was the same for both enzymes and involved first diluting the sample 1:1 with 2,2,2-trifluoroethanol, then reducing with DTT at 90°C for 20 ...
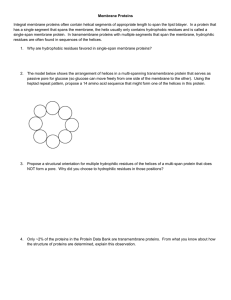
Membrane Proteins Integral membrane proteins often contain
... Integral membrane proteins often contain helical segments of appropriate length to span the lipid bilayer. In a protein that has a single segment that spans the membrane, the helix usually only contains hydrophobic residues and is called a single-span membrane protein. In transmembrane proteins with ...
... Integral membrane proteins often contain helical segments of appropriate length to span the lipid bilayer. In a protein that has a single segment that spans the membrane, the helix usually only contains hydrophobic residues and is called a single-span membrane protein. In transmembrane proteins with ...
Name: TF Name:
... proteins with masses of 19 kDa and 87 kDa is treated with different concentrations of protease. Each lane represents an experiment in which the concentration of the 19 kDa and 87 kDa proteins is constant, but a different concentration of protease was added. Label the sizes of the protein bands in ea ...
... proteins with masses of 19 kDa and 87 kDa is treated with different concentrations of protease. Each lane represents an experiment in which the concentration of the 19 kDa and 87 kDa proteins is constant, but a different concentration of protease was added. Label the sizes of the protein bands in ea ...
Describe the relationship between genes, nucleic acids, amino
... Describe the relationship between genes, nucleic acids, amino acids, and proteins. DNA is a specific nucleic acid that directs protein making in all living things Proteins function in a variety of critical ways in living things not the least of which is to serve as enzymes that catalyze numerous and ...
... Describe the relationship between genes, nucleic acids, amino acids, and proteins. DNA is a specific nucleic acid that directs protein making in all living things Proteins function in a variety of critical ways in living things not the least of which is to serve as enzymes that catalyze numerous and ...
A Database of Peak Annotations of Empirically Derived Mass Spectra
... Presently, several databases containing empirically derived tandem mass spectrum (MS/MS) data are publicly available. These can be used singly or in a concatenated fashion; together they contain the sequences of more than 12 million proteins. We have imported these into the Illinois Bio-Grid Mass Sp ...
... Presently, several databases containing empirically derived tandem mass spectrum (MS/MS) data are publicly available. These can be used singly or in a concatenated fashion; together they contain the sequences of more than 12 million proteins. We have imported these into the Illinois Bio-Grid Mass Sp ...
Introductory Signposting - Oklahoma State University
... not built from experimental mass spec data! ...
... not built from experimental mass spec data! ...
Protein mass spectrometry

Protein mass spectrometry refers to the application of mass spectrometry to the study of proteins. Mass spectrometry is an important emerging method for the characterization of proteins. The two primary methods for ionization of whole proteins are electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI). In keeping with the performance and mass range of available mass spectrometers, two approaches are used for characterizing proteins. In the first, intact proteins are ionized by either of the two techniques described above, and then introduced to a mass analyzer. This approach is referred to as ""top-down"" strategy of protein analysis. In the second, proteins are enzymatically digested into smaller peptides using a protease such as trypsin. Subsequently these peptides are introduced into the mass spectrometer and identified by peptide mass fingerprinting or tandem mass spectrometry. Hence, this latter approach (also called ""bottom-up"" proteomics) uses identification at the peptide level to infer the existence of proteins.Whole protein mass analysis is primarily conducted using either time-of-flight (TOF) MS, or Fourier transform ion cyclotron resonance (FT-ICR). These two types of instrument are preferable here because of their wide mass range, and in the case of FT-ICR, its high mass accuracy. Mass analysis of proteolytic peptides is a much more popular method of protein characterization, as cheaper instrument designs can be used for characterization. Additionally, sample preparation is easier once whole proteins have been digested into smaller peptide fragments. The most widely used instrument for peptide mass analysis are the MALDI time-of-flight instruments as they permit the acquisition of peptide mass fingerprints (PMFs) at high pace (1 PMF can be analyzed in approx. 10 sec). Multiple stage quadrupole-time-of-flight and the quadrupole ion trap also find use in this application.