Mutator Transposon in Maize and MULEs in the Plant Genome
... transposase, which was used as evidence for its very early origin [13]. The mudrB major transcript encodes a 23 kDa protein (MURB) that is not similar to any sequences in public database outside maize and its close relatives. Although the precise function of MURB remains enigmatic, both deletion der ...
... transposase, which was used as evidence for its very early origin [13]. The mudrB major transcript encodes a 23 kDa protein (MURB) that is not similar to any sequences in public database outside maize and its close relatives. Although the precise function of MURB remains enigmatic, both deletion der ...
Factors Affecting synonymous codon Usage Bias in chloroplast
... plant nuclear genes and in chloroplasts.32 Chloroplasts differ from the nuclear genome of higher plants in that they encode only 30 tRNA species. Since chloroplasts have restricted their tRNA genes, the use of preferred codons by chloroplast encoded proteins appears more extreme. However, a positive ...
... plant nuclear genes and in chloroplasts.32 Chloroplasts differ from the nuclear genome of higher plants in that they encode only 30 tRNA species. Since chloroplasts have restricted their tRNA genes, the use of preferred codons by chloroplast encoded proteins appears more extreme. However, a positive ...
software development and application in bioinformatics: single
... d. MAQ aligner: MAQ stands for Mapping and Assembly with Quality. It is a commonly used linux application for short reads alignment and SNP calling [11]. MAQ supports Illumina reads, and includes functions that make it able to handle next-generation sequencing data [11] and AB SOLiD (a parallel next ...
... d. MAQ aligner: MAQ stands for Mapping and Assembly with Quality. It is a commonly used linux application for short reads alignment and SNP calling [11]. MAQ supports Illumina reads, and includes functions that make it able to handle next-generation sequencing data [11] and AB SOLiD (a parallel next ...
Complete Sequence of the Mitochondrial DNA of
... of these genes are encoded in one strand, which is called the ‘‘major strand’’ here, according to Hatzoglou, George, and Lecanidou (1995), and the remaining 13 genes are encoded in the opposite strand, the ‘‘minor strand.’’ One of the remarkable features of Pupa mtDNA is the highly compact genome or ...
... of these genes are encoded in one strand, which is called the ‘‘major strand’’ here, according to Hatzoglou, George, and Lecanidou (1995), and the remaining 13 genes are encoded in the opposite strand, the ‘‘minor strand.’’ One of the remarkable features of Pupa mtDNA is the highly compact genome or ...
Evolution of Immunoglobulin Kappa Chain Variable Region
... a sequence similarity of 95%–100%, and this duplication has not been found in the chimpanzee, gorilla, or orangutan (Ermert et al. 1995). From information on the overall sequence divergence between the two sets of duplicate genes (;1%), Schäble and Zachau (1993) suggested that the duplication occur ...
... a sequence similarity of 95%–100%, and this duplication has not been found in the chimpanzee, gorilla, or orangutan (Ermert et al. 1995). From information on the overall sequence divergence between the two sets of duplicate genes (;1%), Schäble and Zachau (1993) suggested that the duplication occur ...
Genome duplications and accelerated evolution of
... the experimental approaches used to collect the DNA sequences and therefore do not always provide the complete and reliable information that is required for a comprehensive comparative analysis of Hox cluster variation. The most common method employed for the identification of Hox genes has been the ...
... the experimental approaches used to collect the DNA sequences and therefore do not always provide the complete and reliable information that is required for a comprehensive comparative analysis of Hox cluster variation. The most common method employed for the identification of Hox genes has been the ...
publication
... Informatics database map and the Davis Human/Mouse homology map have been made by identifying human orthologs to mapped mouse genes (8). Detailed characterizations of disease regions are increasingly being done through mouse–human genomic comparisons aimed at identifying novel genes and regulatory e ...
... Informatics database map and the Davis Human/Mouse homology map have been made by identifying human orthologs to mapped mouse genes (8). Detailed characterizations of disease regions are increasingly being done through mouse–human genomic comparisons aimed at identifying novel genes and regulatory e ...
Plant and Soil
... and of preparing bacterial isolates that is required for other detection techniques. Using conventional methods only a percentage of the nodules is analyzed, data produced by this method are obtained from the total nodule number. This is an advantage as it is obvious that a larger sample size will s ...
... and of preparing bacterial isolates that is required for other detection techniques. Using conventional methods only a percentage of the nodules is analyzed, data produced by this method are obtained from the total nodule number. This is an advantage as it is obvious that a larger sample size will s ...
Gene prediction and Genome Annotation
... RNA seq issues • RNA seq reads are often very noisy, with reads aligning allover the genome • RNA seq detect sense and antisense transcripts, protein-encoding or not • Only stranded reads can tell for sure in which sense they should apply • Small read length of RNA seq leads to virtual transcripts, ...
... RNA seq issues • RNA seq reads are often very noisy, with reads aligning allover the genome • RNA seq detect sense and antisense transcripts, protein-encoding or not • Only stranded reads can tell for sure in which sense they should apply • Small read length of RNA seq leads to virtual transcripts, ...
Cloning and characterisation of a cysteine proteinase gene
... ORF 1. Considering that the Ldccys2 gene from L. (L.) chagasi also presents a C-terminal extension in its 30 UTR (Omara-Opyene and Gedamu, 1997), besides the high identity between this gene and Llacys1 (88%), the nucleotide sequence of the Ldccys2 30 UTR was compared with those from 3A4 and 2A1 clon ...
... ORF 1. Considering that the Ldccys2 gene from L. (L.) chagasi also presents a C-terminal extension in its 30 UTR (Omara-Opyene and Gedamu, 1997), besides the high identity between this gene and Llacys1 (88%), the nucleotide sequence of the Ldccys2 30 UTR was compared with those from 3A4 and 2A1 clon ...
Comprehensive and Rapid Genotyping of Mutations - HAL
... reported so far are point mutations altering a few bases or only one base (http://www.genet.sickkids.on.ca), while an unknown proportion of CFTR dysfunctions are caused by large genomic rearrangements such as large deletions [1-7]. In CBAVD, 88% of patients found with two CFTR mutations carry a seve ...
... reported so far are point mutations altering a few bases or only one base (http://www.genet.sickkids.on.ca), while an unknown proportion of CFTR dysfunctions are caused by large genomic rearrangements such as large deletions [1-7]. In CBAVD, 88% of patients found with two CFTR mutations carry a seve ...
BROWSING GENES AND GENOMES WITH ENSEMBL
... (c) Have a look at the external references for ENST00000218099. What is the function of F9? (d) Is it possible to monitor expression of ENST00000218099 with the ILLUMINA HumanWG_6_V2 microarray? If so, can it also be used to monitor expression of the other two transcripts? (e) In which part (i.e. th ...
... (c) Have a look at the external references for ENST00000218099. What is the function of F9? (d) Is it possible to monitor expression of ENST00000218099 with the ILLUMINA HumanWG_6_V2 microarray? If so, can it also be used to monitor expression of the other two transcripts? (e) In which part (i.e. th ...
XSL Formatter - H:\XML
... showing that STS-X69962 appears on the STS map. Selecting the STS link gives us a Map Viewer display of the STS we found but not of the gene we sought. To get the gene into the Map Viewer display, we can add the Genes_seq map to the display. Although FMR1 is located on the Genes_seq map rather than ...
... showing that STS-X69962 appears on the STS map. Selecting the STS link gives us a Map Viewer display of the STS we found but not of the gene we sought. To get the gene into the Map Viewer display, we can add the Genes_seq map to the display. Although FMR1 is located on the Genes_seq map rather than ...
MAMMALS THAT BREAK THE RULES:Genetics of Marsupials and
... been made using in situ hybridization with human probes, or cloned wallaby or platypus homologues to conserved human genes. Several genes clustered on human chromosomes have been located together on marsupial and monotreme chromosomes. For instance, three human chromosome (HSA) 11p genes were found ...
... been made using in situ hybridization with human probes, or cloned wallaby or platypus homologues to conserved human genes. Several genes clustered on human chromosomes have been located together on marsupial and monotreme chromosomes. For instance, three human chromosome (HSA) 11p genes were found ...
Applied and Environmental Microbiology
... be the result of gene duplication. In the case of the R. tropici citrate synthase genes, the duplication seems to be ancient since the DNA sequence has clearly diverged outside the coding region, while there is a high degree of similarity in the coding regions of both genes. Besides duplication, the ...
... be the result of gene duplication. In the case of the R. tropici citrate synthase genes, the duplication seems to be ancient since the DNA sequence has clearly diverged outside the coding region, while there is a high degree of similarity in the coding regions of both genes. Besides duplication, the ...
The hybrid origins of three perennial Medicago species
... animal kingdom. Species formed by hybridization often have different phenotype and better fitness than either the parents (Otto 2007). However, in other cases, depending on the relatedness of the parents, the descendants may also be sterile. This is caused by chromosomal ...
... animal kingdom. Species formed by hybridization often have different phenotype and better fitness than either the parents (Otto 2007). However, in other cases, depending on the relatedness of the parents, the descendants may also be sterile. This is caused by chromosomal ...
Meiosis/Crossing Over - Peoria Public Schools
... 10.1.U2 Crossing over is the exchange of DNA material between non-sister homologous chromatids. AND 10.1.U4 Chiasmata formation between non-sister chromatids can result in an exchange of alleles. ...
... 10.1.U2 Crossing over is the exchange of DNA material between non-sister homologous chromatids. AND 10.1.U4 Chiasmata formation between non-sister chromatids can result in an exchange of alleles. ...
Background Selection in Single Genes May Explain
... that the mean allele frequency over the distribution generated by selection, mutation, and drift is well approximated by Equation 1, assuming semidominant effects of mutations on fitness (McVean and Charlesworth 1999). Thus the mean frequency over a group of variants subject to selection is given by ...
... that the mean allele frequency over the distribution generated by selection, mutation, and drift is well approximated by Equation 1, assuming semidominant effects of mutations on fitness (McVean and Charlesworth 1999). Thus the mean frequency over a group of variants subject to selection is given by ...
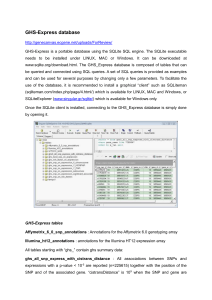
GHS-Express database http://genecanvas.ecgene.net/uploads/Fo
... expressions with a p-value < 10-5 are reported (n=225615) together with the position of the SNP and of the associated gene. “cistransDistance” is 109 when the SNP and gene are ...
... expressions with a p-value < 10-5 are reported (n=225615) together with the position of the SNP and of the associated gene. “cistransDistance” is 109 when the SNP and gene are ...
DNA sequence of the rat growth hormone gene: location of the 5
... All d i g e s t i o n s v/ere done with enzymes purchased fran e i t h e r Mew England Biolabs, rtethesda Pesearch Laboratories, o r Poehringer f'annheim. Digestions were usually done with a s u b s t a n t i a l excess of enzyme and approximately in accordance with t h e conditions provided by t h ...
... All d i g e s t i o n s v/ere done with enzymes purchased fran e i t h e r Mew England Biolabs, rtethesda Pesearch Laboratories, o r Poehringer f'annheim. Digestions were usually done with a s u b s t a n t i a l excess of enzyme and approximately in accordance with t h e conditions provided by t h ...
annotation and analysis of newly discovered mycobacteriophage
... characterize their phage, and then one or more is sequenced by the class. During the course of this project, many mycobacteriophage genomes have been characterized and added to existing databases; this success has driven the production of a mycobacteriophage-specific database that contains all the m ...
... characterize their phage, and then one or more is sequenced by the class. During the course of this project, many mycobacteriophage genomes have been characterized and added to existing databases; this success has driven the production of a mycobacteriophage-specific database that contains all the m ...
Use of QTL analysis in physiological research
... A complication of QTL cloning compared to positional cloning of mutants is finding the gene(s) responsible for the phenotype among the candidate genes in the region where the QTL was fine-mapped. In case of mutants in a self-pollinating species such as Arabidopsis, any DNA sequence difference that i ...
... A complication of QTL cloning compared to positional cloning of mutants is finding the gene(s) responsible for the phenotype among the candidate genes in the region where the QTL was fine-mapped. In case of mutants in a self-pollinating species such as Arabidopsis, any DNA sequence difference that i ...
Plant Genome Resources at the National Center for Biotechnology
... 1995). Locus nomenclature is organism specific and is unreliable as a query method between species; however, the regular nomenclature of plasmids (Lederberg, 1986) is not influenced by how the plasmid or insert is used. The data for the plant maps available through Map Viewer include the marker-locu ...
... 1995). Locus nomenclature is organism specific and is unreliable as a query method between species; however, the regular nomenclature of plasmids (Lederberg, 1986) is not influenced by how the plasmid or insert is used. The data for the plant maps available through Map Viewer include the marker-locu ...
TEL Gene Is Involved in Myelodysplastic Syndromes
... A 1,241-bp TEL cDNA was obtained containing the complete coding sequence. The sequence of this cDNA was identical to the published one.I3 The cDNA was used to screen a chromosome 12 cosmid library. Ten cosmids were obtained and characterized by Southern hybridization with restriction fragments of th ...
... A 1,241-bp TEL cDNA was obtained containing the complete coding sequence. The sequence of this cDNA was identical to the published one.I3 The cDNA was used to screen a chromosome 12 cosmid library. Ten cosmids were obtained and characterized by Southern hybridization with restriction fragments of th ...
Copy-number variation

Copy-number variations (CNVs)—a form of structural variation—are alterations of the DNA of a genome that results in the cell having an abnormal or, for certain genes, a normal variation in the number of copies of one or more sections of the DNA. CNVs correspond to relatively large regions of the genome that have been deleted (fewer than the normal number) or duplicated (more than the normal number) on certain chromosomes. For example, the chromosome that normally has sections in order as A-B-C-D might instead have sections A-B-C-C-D (a duplication of ""C"") or A-B-D (a deletion of ""C"").This variation accounts for roughly 13% of human genomic DNA and each variation may range from about one kilobase (1,000 nucleotide bases) to several megabases in size. CNVs contrast with single-nucleotide polymorphisms (SNPs), which affect only one single nucleotide base.