Supporting information This document contains a comparison
... Among these interactions, H-bonds are the strongest ones. So if we just consider the number of H-bonds observed in these structures we can have a very rough estimate of how much stronger the Arg/PO43- interaction is compared with Glu/ PO43-. Within these structures, the number of Hbonds observed bet ...
... Among these interactions, H-bonds are the strongest ones. So if we just consider the number of H-bonds observed in these structures we can have a very rough estimate of how much stronger the Arg/PO43- interaction is compared with Glu/ PO43-. Within these structures, the number of Hbonds observed bet ...
Answer Key
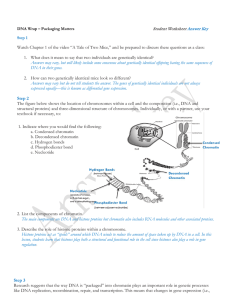
... Answers may vary, but will likely include some consensus about genetically identical offspring having the same sequences of DNA in their genes. 2. How can two genetically identical mice look so different? Answers may vary but do not tell students the answer. The genes of genetically identical indivi ...
... Answers may vary, but will likely include some consensus about genetically identical offspring having the same sequences of DNA in their genes. 2. How can two genetically identical mice look so different? Answers may vary but do not tell students the answer. The genes of genetically identical indivi ...
figure 9-9
... largest types of DNA library is a genomic library, produced when the complete genome of a particular organism is cleaved into thousands of fragments, and all the fragments are cloned by insertion into a cloning vector. 歐亞書局 ...
... largest types of DNA library is a genomic library, produced when the complete genome of a particular organism is cleaved into thousands of fragments, and all the fragments are cloned by insertion into a cloning vector. 歐亞書局 ...
Human Genome Project and Sequencing
... Humans are 99.9% identical. Total number of genes ~ 30,000. This doesn’t match the number of proteins (over 100,000) so each gene must be able to code for more than one protein. Over 50% of genes have unknown ...
... Humans are 99.9% identical. Total number of genes ~ 30,000. This doesn’t match the number of proteins (over 100,000) so each gene must be able to code for more than one protein. Over 50% of genes have unknown ...
Chapter 2 - CSUB Home Page
... 4. The two alleles for each character segregate (separate) during ...
... 4. The two alleles for each character segregate (separate) during ...
DNA
... Due to the various properties of the amino acid side chains, certain residues are found more often in one or the other structural units. Some residues have been classified, for example, as α-helix breakers. Proline for example is a poor helix former due to the fact that its backbone N atom is alread ...
... Due to the various properties of the amino acid side chains, certain residues are found more often in one or the other structural units. Some residues have been classified, for example, as α-helix breakers. Proline for example is a poor helix former due to the fact that its backbone N atom is alread ...
Unit 1 - Calderglen High School
... base/nucleotide is inserted or deleted from a gene (DNA), every codon after where the mutation has occurred will be altered during transcription. This in turn, alters every amino acid after the mutation site. The protein formed is almost certain to be non-functional – in other words the protein will ...
... base/nucleotide is inserted or deleted from a gene (DNA), every codon after where the mutation has occurred will be altered during transcription. This in turn, alters every amino acid after the mutation site. The protein formed is almost certain to be non-functional – in other words the protein will ...
Lab 3 minipreps, RE, gel
... proteins that recognize and bind to specific DNA sequences and cut the DNA at or near the recognition site. Restriction enzymes were originally discovered through their ability to break down, or "restrict" foreign DNA. In their natural environment, the bacterial cell, they serve a protective functio ...
... proteins that recognize and bind to specific DNA sequences and cut the DNA at or near the recognition site. Restriction enzymes were originally discovered through their ability to break down, or "restrict" foreign DNA. In their natural environment, the bacterial cell, they serve a protective functio ...
Virus - Perry Local Schools
... • Both made from RNA molecule that is diced into double stranded (ds) segments. ...
... • Both made from RNA molecule that is diced into double stranded (ds) segments. ...
DNA WebQuest
... Click on “What is DNA?” at the top and go through the animation. Answer the questions. 1) What is DNA? 2) The complete set of instructions for making a human being is found where? 3) What do genes tell the cell to make? Click on “What is a gene?” at the top and go through the animation. Answer the q ...
... Click on “What is DNA?” at the top and go through the animation. Answer the questions. 1) What is DNA? 2) The complete set of instructions for making a human being is found where? 3) What do genes tell the cell to make? Click on “What is a gene?” at the top and go through the animation. Answer the q ...
1999
... Features of Life (as discussed in class / listed in textbook / viewed in lab) – what it means to be ‘alive’ – you don’t need to be able to list them, but you should be able to identify and explain with examples ...
... Features of Life (as discussed in class / listed in textbook / viewed in lab) – what it means to be ‘alive’ – you don’t need to be able to list them, but you should be able to identify and explain with examples ...
Viruses - apbio107
... Begin with a plasmid and the gene of interest. Show how the gene is inserted in the plasmid. Show how the plasmid is incorporated into a bacterial cell. Show how the cell copies the plasmid. Show how the cell expresses the gene of interest. ...
... Begin with a plasmid and the gene of interest. Show how the gene is inserted in the plasmid. Show how the plasmid is incorporated into a bacterial cell. Show how the cell copies the plasmid. Show how the cell expresses the gene of interest. ...
Note_on_isolation_and_DNA_extraction_of_rhizobia
... The formation of symbiotically effective legume root nodules by rhizobia is often expected ex gratia by growers in many countries. However, it is clear that isolation of unique rhizobial strains for culture ex situ and then used as an inoculum may be an inexpensive and effective approach to ensure n ...
... The formation of symbiotically effective legume root nodules by rhizobia is often expected ex gratia by growers in many countries. However, it is clear that isolation of unique rhizobial strains for culture ex situ and then used as an inoculum may be an inexpensive and effective approach to ensure n ...
Click here
... proteins, and other cell components. Basic source of nitrogen is amino acid; others can use ammonia directly through the action of enzymes such as glutamate dehydrogenase or glutamine synthetase and glutamate synthase. Inorganic phosphate is the prime source of phosphorus, and almost all bacteria in ...
... proteins, and other cell components. Basic source of nitrogen is amino acid; others can use ammonia directly through the action of enzymes such as glutamate dehydrogenase or glutamine synthetase and glutamate synthase. Inorganic phosphate is the prime source of phosphorus, and almost all bacteria in ...
13-2 Manipulating DNA
... Reading the Sequence Researchers use a clever chemical trick to "read" DNA by determining the order of its bases. A single strand of DNA whose sequence of bases is not known is placed in a test tube. DNA polymerase, the enzyme that copies DNA, and the four nucleotide bases, A, T, G, and C, are added ...
... Reading the Sequence Researchers use a clever chemical trick to "read" DNA by determining the order of its bases. A single strand of DNA whose sequence of bases is not known is placed in a test tube. DNA polymerase, the enzyme that copies DNA, and the four nucleotide bases, A, T, G, and C, are added ...
18 - cloudfront.net
... Reading the Sequence Researchers use a clever chemical trick to "read" DNA by determining the order of its bases. A single strand of DNA whose sequence of bases is not known is placed in a test tube. DNA polymerase, the enzyme that copies DNA, and the four nucleotide bases, A, T, G, and C, are added ...
... Reading the Sequence Researchers use a clever chemical trick to "read" DNA by determining the order of its bases. A single strand of DNA whose sequence of bases is not known is placed in a test tube. DNA polymerase, the enzyme that copies DNA, and the four nucleotide bases, A, T, G, and C, are added ...
Regulation of Gene Expression
... pathway are often found in the same operon – All under the control of the same promoter region – Thus these genes are transcribed all together into one continuous mRNA strand: polycistronic mRNA • Proteins are then synthesized from that mRNA ...
... pathway are often found in the same operon – All under the control of the same promoter region – Thus these genes are transcribed all together into one continuous mRNA strand: polycistronic mRNA • Proteins are then synthesized from that mRNA ...
DNA Technology
... the plasmid (only one recognition site), and human DNA Result- human DNA is cut into many fragments – one is the correct one. Cut plasmids and DNA fragments are mixed ...
... the plasmid (only one recognition site), and human DNA Result- human DNA is cut into many fragments – one is the correct one. Cut plasmids and DNA fragments are mixed ...
The subcomponents of biological molecules and their sequence
... Protein’s property is really dependent on its subcomponents; for protein everything matters. The R group is vital in proteins because the R groups of amino acids are the components that make the final shape of the protein and the function of the protein. The bonding in the tertiary structure is all ...
... Protein’s property is really dependent on its subcomponents; for protein everything matters. The R group is vital in proteins because the R groups of amino acids are the components that make the final shape of the protein and the function of the protein. The bonding in the tertiary structure is all ...
Slide ()
... Comparison of the structure of the human visual pigment genes. Coding sequences of the genes are denoted by boxes and noncoding regions by lines (not to scale). Open boxes represent untranslated regions, and filled boxes denote the coding regions. The length of introns in number of base pairs is sho ...
... Comparison of the structure of the human visual pigment genes. Coding sequences of the genes are denoted by boxes and noncoding regions by lines (not to scale). Open boxes represent untranslated regions, and filled boxes denote the coding regions. The length of introns in number of base pairs is sho ...
Unit 6 ~ Learning Guide Name: INSTRUCTIONS
... 3. Please list the steps of translation and provide a brief description of each step. (6 marks) Step 1. = Initiation where small ribosome subunit binds to start codon of mRNA, the initiator tRNA binds complimentary to the mRNA's start codon and then the large ribosomal subunit binds to the complex s ...
... 3. Please list the steps of translation and provide a brief description of each step. (6 marks) Step 1. = Initiation where small ribosome subunit binds to start codon of mRNA, the initiator tRNA binds complimentary to the mRNA's start codon and then the large ribosomal subunit binds to the complex s ...
Assignment Chapter 12: Aldehydes, Ketones and Carboxylic Acids 1
... An organic compound contains 69.77% carbon, 11.63% hydrogen and the rest is oxygen. The molecular mass of the compound is 86. It does not reduce Tollen’s reagent but forms an addition compound with sodium hydrogen sulphite and gives a positive iodoform test. On vigorous oxidation it gives ethanoic a ...
... An organic compound contains 69.77% carbon, 11.63% hydrogen and the rest is oxygen. The molecular mass of the compound is 86. It does not reduce Tollen’s reagent but forms an addition compound with sodium hydrogen sulphite and gives a positive iodoform test. On vigorous oxidation it gives ethanoic a ...
Archaea are prokaryotic
... So how do thermophilic organisms manage to live and reproduce? It turns out that they have their own versions of these enzymes that are specially equipped to work at high temperatures. In 1976, Chien, Edgar, and Trela isolated a DNA polymerase from T. aquaticus, and found that it had a temperature o ...
... So how do thermophilic organisms manage to live and reproduce? It turns out that they have their own versions of these enzymes that are specially equipped to work at high temperatures. In 1976, Chien, Edgar, and Trela isolated a DNA polymerase from T. aquaticus, and found that it had a temperature o ...
Nucleic acid analogue
Nucleic acid analogues are compounds which are analogous (structurally similar) to naturally occurring RNA and DNA, used in medicine and in molecular biology research.Nucleic acids are chains of nucleotides, which are composed of three parts: a phosphate backbone, a pucker-shaped pentose sugar, either ribose or deoxyribose, and one of four nucleobases.An analogue may have any of these altered. Typically the analogue nucleobases confer, among other things, different base pairing and base stacking properties. Examples include universal bases, which can pair with all four canonical bases, and phosphate-sugar backbone analogues such as PNA, which affect the properties of the chain (PNA can even form a triple helix).Nucleic acid analogues are also called Xeno Nucleic Acid and represent one of the main pillars of xenobiology, the design of new-to-nature forms of life based on alternative biochemistries.Artificial nucleic acids include peptide nucleic acid (PNA), Morpholino and locked nucleic acid (LNA), as well as glycol nucleic acid (GNA) and threose nucleic acid (TNA). Each of these is distinguished from naturally occurring DNA or RNA by changes to the backbone of the molecule.In May 2014, researchers announced that they had successfully introduced two new artificial nucleotides into bacterial DNA, and by including individual artificial nucleotides in the culture media, were able to passage the bacteria 24 times; they did not create mRNA or proteins able to use the artificial nucleotides. The artificial nucleotides featured 2 fused aromatic rings.