Lecture 1
... An overview of the mechanisms that can be used in regulation.The product of gene A is an enzyme A, which in this case is synthesised constitutively and carries out its reaction. Enzyme B is also synthesised constitutively but its activity can be inhibited. The synthesis of the product of gene C can ...
... An overview of the mechanisms that can be used in regulation.The product of gene A is an enzyme A, which in this case is synthesised constitutively and carries out its reaction. Enzyme B is also synthesised constitutively but its activity can be inhibited. The synthesis of the product of gene C can ...
Computer science
... “We can approach understanding how the whole genome works by breaking it down into groups of genes that interact strongly with each other. Once researchers identify and understand these network modules, the next step will be to figure out the interactions within networks of networks, and so on until ...
... “We can approach understanding how the whole genome works by breaking it down into groups of genes that interact strongly with each other. Once researchers identify and understand these network modules, the next step will be to figure out the interactions within networks of networks, and so on until ...
Characterization of Genes Expressed During the Early Stages of
... Subtractive cloning is a powerful method for detecting and isolating gene sequences that are differentially expressed. This approach was used to isolate genes expressed during the early induction period in the alfalfa direct somatic embryogenic system, and nearly 100 different clones were identified ...
... Subtractive cloning is a powerful method for detecting and isolating gene sequences that are differentially expressed. This approach was used to isolate genes expressed during the early induction period in the alfalfa direct somatic embryogenic system, and nearly 100 different clones were identified ...
Tensor Decomposition of Microarray Data - DIMACS REU
... The support of the Rutgers University DIMACS REU is gratefully acknowledged. ...
... The support of the Rutgers University DIMACS REU is gratefully acknowledged. ...
Yeast Cell-Cycle Regulation Network inference
... • Problem1: Cluster using the whole data set always did not show good results because of the noise and the complex network. • Problem2: How to select a cluster method. ...
... • Problem1: Cluster using the whole data set always did not show good results because of the noise and the complex network. • Problem2: How to select a cluster method. ...
Note 7.4 - Controlling Gene Expression
... Posttranslational: before many proteins become functional, they must pass through the cell membrane. A number of control mechanisms affect the rate at which a protein becomes active and the time it remains functional, including the addition of various chemical groups. ...
... Posttranslational: before many proteins become functional, they must pass through the cell membrane. A number of control mechanisms affect the rate at which a protein becomes active and the time it remains functional, including the addition of various chemical groups. ...
differential gene expression
... Multicellular Organisms • Almost all the cells in an organism are genetically identical or totipotent. • Differences between cell types result from differential gene expression -- the expression of different genes by cells with the same genome. • Errors in gene expression can lead to diseases includ ...
... Multicellular Organisms • Almost all the cells in an organism are genetically identical or totipotent. • Differences between cell types result from differential gene expression -- the expression of different genes by cells with the same genome. • Errors in gene expression can lead to diseases includ ...
Gene Expression and Regulation
... • Similar process, but is much more complex than prokaryotic gene regulation – Prokaryotes have no cell specialization ...
... • Similar process, but is much more complex than prokaryotic gene regulation – Prokaryotes have no cell specialization ...
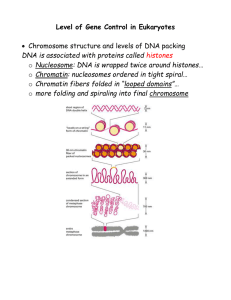
Genetic Controls in Eukaryotes
... o “General” transcription factors leads to slow transcription. - General = essential to initiation of transcription of all protein o “Specific” transcription factors leads to faster transcription = Specific to transcription of particular protein. ...
... o “General” transcription factors leads to slow transcription. - General = essential to initiation of transcription of all protein o “Specific” transcription factors leads to faster transcription = Specific to transcription of particular protein. ...
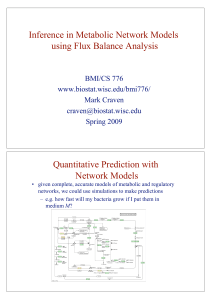
Inference in Metabolic Network Models using Flux
... •! the time constants associated with transcriptional regulation (minutes) and cell growth (hours) are slow •! quasi steady-state assumption: behavior inside cell is in steadystate during short time intervals •! can do simulations by iteratively •! changing representation of external environment (e. ...
... •! the time constants associated with transcriptional regulation (minutes) and cell growth (hours) are slow •! quasi steady-state assumption: behavior inside cell is in steadystate during short time intervals •! can do simulations by iteratively •! changing representation of external environment (e. ...
Checklist unit 18: Regulation of Gene Expression
... prokaryotic and eukaryotic cell. Gene regulation is crucial for organism development, responses to environmental changes, and the allocation of metabolic resources. Organisms must be able to turn protein synthesis on and off; this is accomplished through the regulation of gene expression. As we lear ...
... prokaryotic and eukaryotic cell. Gene regulation is crucial for organism development, responses to environmental changes, and the allocation of metabolic resources. Organisms must be able to turn protein synthesis on and off; this is accomplished through the regulation of gene expression. As we lear ...
Coarse-Graining of Macromolecules
... For us, the whole question of transcriptional regulation will come down to the question of whether or not RNAP is bound to the promoter or not! There are an array of molecules (transcription factors) that participate in recruiting RNAP to its promoter. (Ptashne and Gann) ...
... For us, the whole question of transcriptional regulation will come down to the question of whether or not RNAP is bound to the promoter or not! There are an array of molecules (transcription factors) that participate in recruiting RNAP to its promoter. (Ptashne and Gann) ...
MTC19: transcription and gene expression 02/10/07
... A gene in transcription can be defined as a segment of DNA extending from the site of initiation to the site of termination Genes consist of exons (sequences to be subsequently translated into proteins) separated by introns, which can contain other control regions or even other genes to allow more c ...
... A gene in transcription can be defined as a segment of DNA extending from the site of initiation to the site of termination Genes consist of exons (sequences to be subsequently translated into proteins) separated by introns, which can contain other control regions or even other genes to allow more c ...
Media:RuthNov07pres
... • Approach framework will be to quantitatively monitor genome-wide RNA and protein levels in a model DHC strain (D. ethenogenes strain 195 - DET) growing in mixed-culture conditions in pseudo-steady-state reactors and to utilize systems biology algorithms of network inference to compile the data int ...
... • Approach framework will be to quantitatively monitor genome-wide RNA and protein levels in a model DHC strain (D. ethenogenes strain 195 - DET) growing in mixed-culture conditions in pseudo-steady-state reactors and to utilize systems biology algorithms of network inference to compile the data int ...
Gene Regulation
... Cells regulate what genes are expressed based on what protein the cell NEEDS. ...
... Cells regulate what genes are expressed based on what protein the cell NEEDS. ...
Brief description of pGLO
... Regulation of the genes for arabinose metabolism provides an interesting comparison to regulation of the lac operon. (Review the previous diagram of arabinose metabolism in E. coli.) The 3 genes for arabinose metabolism; araB , araA, and araD are transcribed, in that order, onto a single polycistron ...
... Regulation of the genes for arabinose metabolism provides an interesting comparison to regulation of the lac operon. (Review the previous diagram of arabinose metabolism in E. coli.) The 3 genes for arabinose metabolism; araB , araA, and araD are transcribed, in that order, onto a single polycistron ...
In-vivo and in-vitro investigation of Aspirin using pan
... identified which showed more than 20-fold changes. Altered gene activities comprise e.g. PI3 a gene encoding an elastase-specific inhibitor or MMP7, a gene encoding Proteins of the matrix metalloproteinase (MMP) family involved in the breakdown of extracellular matrix. Four h after i.v. administrati ...
... identified which showed more than 20-fold changes. Altered gene activities comprise e.g. PI3 a gene encoding an elastase-specific inhibitor or MMP7, a gene encoding Proteins of the matrix metalloproteinase (MMP) family involved in the breakdown of extracellular matrix. Four h after i.v. administrati ...
Genetic Control - Deans Community High School
... Essential proteins (for enzymes and structure) are synthesised according to the base sequence encoded in the cell’s DNA. A particular segment of DNA called a gene codes for each protein. Thus the structure and function of cell is determined and controlled by its genes. All undifferentiated cells hav ...
... Essential proteins (for enzymes and structure) are synthesised according to the base sequence encoded in the cell’s DNA. A particular segment of DNA called a gene codes for each protein. Thus the structure and function of cell is determined and controlled by its genes. All undifferentiated cells hav ...
file1 - Department of Computer Science
... Model more complex organisms Genetic engineering Drugs ...
... Model more complex organisms Genetic engineering Drugs ...
Regulation of Gene Expression
... • Genes that are involved in the same metabolic pathway are often found in the same operon – All under the control of the same promoter region – Thus these genes are transcribed all together into one continuous mRNA strand: polycistronic mRNA • Proteins are then synthesized from that mRNA ...
... • Genes that are involved in the same metabolic pathway are often found in the same operon – All under the control of the same promoter region – Thus these genes are transcribed all together into one continuous mRNA strand: polycistronic mRNA • Proteins are then synthesized from that mRNA ...
Chapter 10 Section 3 Notes Answer Key
... I. DNA A. DNA- a chemical that contains information an organism needs to grow and function 1. Watson and Crick made and accurate model of DNA in 1953 2. The structure of DNA is similar to a twisted ladder. a. The sides of the ladder are made up of sugarphosphate molecules. b. The rungs of the ladder ...
... I. DNA A. DNA- a chemical that contains information an organism needs to grow and function 1. Watson and Crick made and accurate model of DNA in 1953 2. The structure of DNA is similar to a twisted ladder. a. The sides of the ladder are made up of sugarphosphate molecules. b. The rungs of the ladder ...
The aim of the thesis was to characterize chosen expression vectors
... their use in studies of promoter activity control by sigma factors of RNA polymerase. Different properties of these vectors (level of expression of the cloned gene, leaky expression without inducer, dependence of expression level on inducer concentration and cell population homogeneity) were found b ...
... their use in studies of promoter activity control by sigma factors of RNA polymerase. Different properties of these vectors (level of expression of the cloned gene, leaky expression without inducer, dependence of expression level on inducer concentration and cell population homogeneity) were found b ...
Dynamic Network Inference
... Dynamic Network Inference Most statistical work is done on gene regulatory networks, while inference of metabolic pathways and signaling networks are done by other means. Like in phylogenetics, network inference has two components – graph structure (topology) and continuous aspects, such as paramete ...
... Dynamic Network Inference Most statistical work is done on gene regulatory networks, while inference of metabolic pathways and signaling networks are done by other means. Like in phylogenetics, network inference has two components – graph structure (topology) and continuous aspects, such as paramete ...
Gene regulatory network

A gene regulatory network or genetic regulatory network (GRN) is a collection of regulators thatinteract with each other and with other substances in the cell to govern the gene expression levels of mRNA and proteins.The regulator can be DNA, RNA, protein and their complex. The interaction can be direct or indirect (through their transcribed RNA or translated protein).In general, each mRNA molecule goes on to make a specific protein (or set of proteins). In some cases this protein will be structural, and will accumulate at the cell membrane or within the cell to give it particular structural properties. In other cases the protein will be an enzyme, i.e., a micro-machine that catalyses a certain reaction, such as the breakdown of a food source or toxin. Some proteins though serve only to activate other genes, and these are the transcription factors that are the main players in regulatory networks or cascades. By binding to the promoter region at the start of other genes they turn them on, initiating the production of another protein, and so on. Some transcription factors are inhibitory.In single-celled organisms, regulatory networks respond to the external environment, optimising the cell at a given time for survival in this environment. Thus a yeast cell, finding itself in a sugar solution, will turn on genes to make enzymes that process the sugar to alcohol. This process, which we associate with wine-making, is how the yeast cell makes its living, gaining energy to multiply, which under normal circumstances would enhance its survival prospects.In multicellular animals the same principle has been put in the service of gene cascades that control body-shape. Each time a cell divides, two cells result which, although they contain the same genome in full, can differ in which genes are turned on and making proteins. Sometimes a 'self-sustaining feedback loop' ensures that a cell maintains its identity and passes it on. Less understood is the mechanism of epigenetics by which chromatin modification may provide cellular memory by blocking or allowing transcription. A major feature of multicellular animals is the use of morphogen gradients, which in effect provide a positioning system that tells a cell where in the body it is, and hence what sort of cell to become. A gene that is turned on in one cell may make a product that leaves the cell and diffuses through adjacent cells, entering them and turning on genes only when it is present above a certain threshold level. These cells are thus induced into a new fate, and may even generate other morphogens that signal back to the original cell. Over longer distances morphogens may use the active process of signal transduction. Such signalling controls embryogenesis, the building of a body plan from scratch through a series of sequential steps. They also control and maintain adult bodies through feedback processes, and the loss of such feedback because of a mutation can be responsible for the cell proliferation that is seen in cancer. In parallel with this process of building structure, the gene cascade turns on genes that make structural proteins that give each cell the physical properties it needs.It has been suggested that, because biological molecular interactions are intrinsically stochastic, gene networks are the result of cellular processes and not their cause (i.e. cellular Darwinism). However, recent experimental evidence has favored the attractor view of cell fates.