Differential expression of genes involved in
... Analysis of gene expression data agrees with certain findings of Berney et al. (2010) • Berney et al. (2010) shows up-regulation of NAD+/NADH independent enzymes coupled with down regulation of NAD+/NADH dependent enzymes • Berney et al. (2010) shows differential regulation of ferredoxin usage and ...
... Analysis of gene expression data agrees with certain findings of Berney et al. (2010) • Berney et al. (2010) shows up-regulation of NAD+/NADH independent enzymes coupled with down regulation of NAD+/NADH dependent enzymes • Berney et al. (2010) shows differential regulation of ferredoxin usage and ...
Topic Definition 3` Refers to the third carbon of the nucleic acid
... This mechanism is utilized to generate a series of closely related protein isoforms, which differ by the inclusion or exclusion of the particular protein domains encoded by those exons. Alternative splicing is directed by RNA-binding proteins that block, or stimulate, utilization of a particular spl ...
... This mechanism is utilized to generate a series of closely related protein isoforms, which differ by the inclusion or exclusion of the particular protein domains encoded by those exons. Alternative splicing is directed by RNA-binding proteins that block, or stimulate, utilization of a particular spl ...
Topic Definition 3` Refers to the third carbon of the nucleic acid
... This mechanism is utilized to generate a series of closely related protein isoforms, which differ by the inclusion or exclusion of the particular protein domains encoded by those exons. Alternative splicing is directed by RNA-binding proteins that block, or stimulate, utilization of a particular spl ...
... This mechanism is utilized to generate a series of closely related protein isoforms, which differ by the inclusion or exclusion of the particular protein domains encoded by those exons. Alternative splicing is directed by RNA-binding proteins that block, or stimulate, utilization of a particular spl ...
Extra Chromosomal Elements
... Natural plasmids are present naturally in bacterial and some yeast cells carrying genes for its own replication and genes for some functions of the cells like F- plasmid (F-pili during conjugation and some R-plasmids for drug resistance). Artificial plasmids are naturally present plasmid but designe ...
... Natural plasmids are present naturally in bacterial and some yeast cells carrying genes for its own replication and genes for some functions of the cells like F- plasmid (F-pili during conjugation and some R-plasmids for drug resistance). Artificial plasmids are naturally present plasmid but designe ...
Table of Contents - Milan Area Schools
... Translation: RNA-Directed Polypeptide Synthesis • Translation begins with an initiation complex: a charged tRNA with its amino acid and a small subunit, both bound to the mRNA. ...
... Translation: RNA-Directed Polypeptide Synthesis • Translation begins with an initiation complex: a charged tRNA with its amino acid and a small subunit, both bound to the mRNA. ...
MicroReview Expression, secretion and antigenic - UvA-DARE
... S-protein represents approx. 10 to 15% of the total cellular protein of the bacterial cell. S-protein genes thus must be efficiently transcribed and/or yield stable mRNA. Multiple promoters have been found in front of the S-protein genes of several species, while S-protein genes of other species are ...
... S-protein represents approx. 10 to 15% of the total cellular protein of the bacterial cell. S-protein genes thus must be efficiently transcribed and/or yield stable mRNA. Multiple promoters have been found in front of the S-protein genes of several species, while S-protein genes of other species are ...
Evolution of Metabolic Pathway
... ¾ Surface proteins of bacteria as drug targets ¾ Host interaction factors that are essential for establishing parasitic state. ¾ Analogous proteins in certain pathogens in a different form that is present in humans. Detailed analysis of non-orthologous displacement cases has led to suggestions that ...
... ¾ Surface proteins of bacteria as drug targets ¾ Host interaction factors that are essential for establishing parasitic state. ¾ Analogous proteins in certain pathogens in a different form that is present in humans. Detailed analysis of non-orthologous displacement cases has led to suggestions that ...
when glucose is scarce
... 1. A repressible operon is one that is usually on; binding of a repressor to the operator shuts off transcription. 1. The trp operon is a repressible operon. trp operon ...
... 1. A repressible operon is one that is usually on; binding of a repressor to the operator shuts off transcription. 1. The trp operon is a repressible operon. trp operon ...
Gene Expression
... Key to understanding real-time PCR is the exponential growth of amplicon during the PCR cycling process. An assumption is that the PCR reaction is running at 100% efficient, meaning that there is a complete doubling of amplicon during the geometric phase of amplification. Figure 1 shows an image tak ...
... Key to understanding real-time PCR is the exponential growth of amplicon during the PCR cycling process. An assumption is that the PCR reaction is running at 100% efficient, meaning that there is a complete doubling of amplicon during the geometric phase of amplification. Figure 1 shows an image tak ...
1. Overview of Gene Expression Overview of Gene Expression Chapter 10B:
... • when we talk about “genes” we will focus on those that express proteins ( the “end products” for a small percentage of genes are special types of RNA molecules) ...
... • when we talk about “genes” we will focus on those that express proteins ( the “end products” for a small percentage of genes are special types of RNA molecules) ...
Chapter 1 - Test bank for TextBook
... that is read three nucleotides at a time to direct the building of proteins. 4. Humans may have the same genes but differ genetically in the alleles they carry. 5. Differential gene expression creates the distinctive cell types. 6. The assumption is that changes in DNA sequence accumulate over time. ...
... that is read three nucleotides at a time to direct the building of proteins. 4. Humans may have the same genes but differ genetically in the alleles they carry. 5. Differential gene expression creates the distinctive cell types. 6. The assumption is that changes in DNA sequence accumulate over time. ...
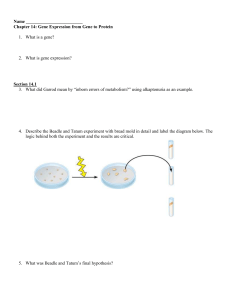
Chapter 14 Guided Reading
... 20. Use the diagram below to understand transcription: Label the diagrams and discuss what is happening. ...
... 20. Use the diagram below to understand transcription: Label the diagrams and discuss what is happening. ...
Distinguish between mRNA, rRNA, and tRNA. What molecule does
... information carried by a gene. mRNA is transcribed from a DNA template, and carries information to the sites of protein synthesis: the ribosome. ...
... information carried by a gene. mRNA is transcribed from a DNA template, and carries information to the sites of protein synthesis: the ribosome. ...
syllabus
... for thiamin, riboflavin, niacin, vitamin B6, folate, vitamin B12, pantothenic acid, biotin, and choline National Academy Press, Washington, D.C. 5. 國人膳食營養素參考攝取量及其說明 行政院衛生署 修訂第六版 92 年 9 月 6. Handouts from NS103 course in the Department of Nutritional Sciences and Toxicology, UC Berkeley ...
... for thiamin, riboflavin, niacin, vitamin B6, folate, vitamin B12, pantothenic acid, biotin, and choline National Academy Press, Washington, D.C. 5. 國人膳食營養素參考攝取量及其說明 行政院衛生署 修訂第六版 92 年 9 月 6. Handouts from NS103 course in the Department of Nutritional Sciences and Toxicology, UC Berkeley ...
Methods S1
... DREB2A fragment spanning amino acid residues 136-335 (DREB2A CT) that was expressed in Escherichia coli as an antigen. The coding sequence of DREB2A CT was amplified by PCR from a cDNA clone of DREB2A [5] using the primer pair DREB2A/406F-EcoRI DREB2A/C-SalI ...
... DREB2A fragment spanning amino acid residues 136-335 (DREB2A CT) that was expressed in Escherichia coli as an antigen. The coding sequence of DREB2A CT was amplified by PCR from a cDNA clone of DREB2A [5] using the primer pair DREB2A/406F-EcoRI DREB2A/C-SalI ...
Studying gene expression with genomic data and Codon Adaptation
... only when a correct dataset is used for “inference” on gene expression the genes are distributed according to a normal-like function which possess a reasonably low average value. In fact, when randomized dataset are used (both genomic randomization and reference set randomization) the genes present ...
... only when a correct dataset is used for “inference” on gene expression the genes are distributed according to a normal-like function which possess a reasonably low average value. In fact, when randomized dataset are used (both genomic randomization and reference set randomization) the genes present ...
... generated from researchers through experiments performed in laboratory across the world. The gene H. sapiens in Entrez Gene (EG), amyloid beta (A4) precursor protein (APP) contain details and comprehensive information about gene and other related elements. Annotation data cross-reference database is ...
Chapter 7 Molecular Genetics: From DNA to Proteins Worksheets
... Read these passages from the text and answer the questions that follow. Beneficial Mutations Some mutations have a positive effect on the organism in which they occur. They are called beneficial mutations. They lead to new versions of proteins that help organisms adapt to changes in their environment. ...
... Read these passages from the text and answer the questions that follow. Beneficial Mutations Some mutations have a positive effect on the organism in which they occur. They are called beneficial mutations. They lead to new versions of proteins that help organisms adapt to changes in their environment. ...
Image PowerPoint
... Cross-over then occurs between non-homologous sections. As a result, genes are duplicated on one chromosome, and deleted on the other. Unequal cross-over events are thought to produce gene duplication in eukaryotic evolution, providing new possibilities for gene function. ...
... Cross-over then occurs between non-homologous sections. As a result, genes are duplicated on one chromosome, and deleted on the other. Unequal cross-over events are thought to produce gene duplication in eukaryotic evolution, providing new possibilities for gene function. ...
An Introduction to Transfection Methods
... of RNA and protein, as well as the applicability to use this technique for both transient and stable protein production. Despite these advantages, there are several drawbacks, including low efficiencies in most primary cells, as well as suspension cell lines, linked to the dependence on endocytotic ...
... of RNA and protein, as well as the applicability to use this technique for both transient and stable protein production. Despite these advantages, there are several drawbacks, including low efficiencies in most primary cells, as well as suspension cell lines, linked to the dependence on endocytotic ...
Title - Iowa State University
... substrate inhibits catalysis by binding to the enzyme’s active site. c. A regulatory molecule binds at a location other than the active site and changes the shape of the enzyme in a way that makes the active site unavailable to the enzyme’s natural substrates. d. Regulatory molecules breakdown carbo ...
... substrate inhibits catalysis by binding to the enzyme’s active site. c. A regulatory molecule binds at a location other than the active site and changes the shape of the enzyme in a way that makes the active site unavailable to the enzyme’s natural substrates. d. Regulatory molecules breakdown carbo ...
Today`s Plan: 4/25/03
... and scientists need a way to check to see if their transgenic bacteria picked up the donor gene. • To test this, an antibiotic resistant gene is often also transferred with the desired donor gene. • We can then grow the bacteria up on a plate containing the antibiotic in the agar. Only resistant bac ...
... and scientists need a way to check to see if their transgenic bacteria picked up the donor gene. • To test this, an antibiotic resistant gene is often also transferred with the desired donor gene. • We can then grow the bacteria up on a plate containing the antibiotic in the agar. Only resistant bac ...
Bio102 Problems
... 22A. Label the 5′ and 3′ ends on the DNA sequence and indicate which one is the template strand. 22B. Label and name both UTRs. 22C. The promoter consists of two key sequences. Name both of them and show where on the DNA or RNA sequence they would be expected. (You do not have to know or find their ...
... 22A. Label the 5′ and 3′ ends on the DNA sequence and indicate which one is the template strand. 22B. Label and name both UTRs. 22C. The promoter consists of two key sequences. Name both of them and show where on the DNA or RNA sequence they would be expected. (You do not have to know or find their ...
Gene regulatory network

A gene regulatory network or genetic regulatory network (GRN) is a collection of regulators thatinteract with each other and with other substances in the cell to govern the gene expression levels of mRNA and proteins.The regulator can be DNA, RNA, protein and their complex. The interaction can be direct or indirect (through their transcribed RNA or translated protein).In general, each mRNA molecule goes on to make a specific protein (or set of proteins). In some cases this protein will be structural, and will accumulate at the cell membrane or within the cell to give it particular structural properties. In other cases the protein will be an enzyme, i.e., a micro-machine that catalyses a certain reaction, such as the breakdown of a food source or toxin. Some proteins though serve only to activate other genes, and these are the transcription factors that are the main players in regulatory networks or cascades. By binding to the promoter region at the start of other genes they turn them on, initiating the production of another protein, and so on. Some transcription factors are inhibitory.In single-celled organisms, regulatory networks respond to the external environment, optimising the cell at a given time for survival in this environment. Thus a yeast cell, finding itself in a sugar solution, will turn on genes to make enzymes that process the sugar to alcohol. This process, which we associate with wine-making, is how the yeast cell makes its living, gaining energy to multiply, which under normal circumstances would enhance its survival prospects.In multicellular animals the same principle has been put in the service of gene cascades that control body-shape. Each time a cell divides, two cells result which, although they contain the same genome in full, can differ in which genes are turned on and making proteins. Sometimes a 'self-sustaining feedback loop' ensures that a cell maintains its identity and passes it on. Less understood is the mechanism of epigenetics by which chromatin modification may provide cellular memory by blocking or allowing transcription. A major feature of multicellular animals is the use of morphogen gradients, which in effect provide a positioning system that tells a cell where in the body it is, and hence what sort of cell to become. A gene that is turned on in one cell may make a product that leaves the cell and diffuses through adjacent cells, entering them and turning on genes only when it is present above a certain threshold level. These cells are thus induced into a new fate, and may even generate other morphogens that signal back to the original cell. Over longer distances morphogens may use the active process of signal transduction. Such signalling controls embryogenesis, the building of a body plan from scratch through a series of sequential steps. They also control and maintain adult bodies through feedback processes, and the loss of such feedback because of a mutation can be responsible for the cell proliferation that is seen in cancer. In parallel with this process of building structure, the gene cascade turns on genes that make structural proteins that give each cell the physical properties it needs.It has been suggested that, because biological molecular interactions are intrinsically stochastic, gene networks are the result of cellular processes and not their cause (i.e. cellular Darwinism). However, recent experimental evidence has favored the attractor view of cell fates.