Site-specific recombination mechanisms exploit DNA
... bacteriophage (Mu) changes its host range through expression of different tail fibers by changing the orientation of a specific DNA segment, the G segment, in its genome1. The phage-encoded Gin recombinase protein specifically recombined the G segment located between short inverted DNA sequences, bu ...
... bacteriophage (Mu) changes its host range through expression of different tail fibers by changing the orientation of a specific DNA segment, the G segment, in its genome1. The phage-encoded Gin recombinase protein specifically recombined the G segment located between short inverted DNA sequences, bu ...
Sequencing a genome - Information Services and Technology
... independently of the chromosomes; artificial plasmids can be inserted into bacteria to amplify DNA for sequencing ...
... independently of the chromosomes; artificial plasmids can be inserted into bacteria to amplify DNA for sequencing ...
Whole genome sequencing - Center for Biological Sequence Analysis
... (capillary Sanger sequencing) ...
... (capillary Sanger sequencing) ...
Genetic Changes = Mutations
... d. structural problems 4. false 5. Similarities: both involve DNA Both might result in either positive or negative Differences: Body cell DNA mutations affect the individual Sex cell DNA mutations affect the next generation 6. cancer … uncontrolled cell division 7. Point mutation: a change in a sing ...
... d. structural problems 4. false 5. Similarities: both involve DNA Both might result in either positive or negative Differences: Body cell DNA mutations affect the individual Sex cell DNA mutations affect the next generation 6. cancer … uncontrolled cell division 7. Point mutation: a change in a sing ...
PowerPoint Presentation - No Slide Title
... Agarose Gel Electrophoresis separates DNA fragments based on their size. DNA fragments are often detected using fluorescence. ...
... Agarose Gel Electrophoresis separates DNA fragments based on their size. DNA fragments are often detected using fluorescence. ...
Clike here - University of Evansville Faculty Web sites
... Agarose Gel Electrophoresis separates DNA fragments based on their size. DNA fragments are often detected using fluorescence. ...
... Agarose Gel Electrophoresis separates DNA fragments based on their size. DNA fragments are often detected using fluorescence. ...
App1PCR - FSU Biology
... APPENDIX 1: THE POLYMERASE CHAIN REACTION The polymerase chain reaction, or PCR, is a technique that allows for the amplification of a specific target DNA sequence within a larger population of DNA (such as the human genome). Using PCR, picogram quantities of target DNA can be amplified to yield mic ...
... APPENDIX 1: THE POLYMERASE CHAIN REACTION The polymerase chain reaction, or PCR, is a technique that allows for the amplification of a specific target DNA sequence within a larger population of DNA (such as the human genome). Using PCR, picogram quantities of target DNA can be amplified to yield mic ...
Supplemental Material
... amplified a 5'- and 3'-flanking regions of chkA and chkB genes and a final fusion PCR with the marker gene. For the DNA fragments containing the 5’- and 3’gene flanking regions, genomic DNA from FGSC A4 strain was used as a template. Table 2 shows the primer sequences and fragment sizes for chkA and ...
... amplified a 5'- and 3'-flanking regions of chkA and chkB genes and a final fusion PCR with the marker gene. For the DNA fragments containing the 5’- and 3’gene flanking regions, genomic DNA from FGSC A4 strain was used as a template. Table 2 shows the primer sequences and fragment sizes for chkA and ...
Who wants to be a millionaire template
... Be coded on hemoglobin, but for Sickle-cell Anemia this amino acid is coded for ...
... Be coded on hemoglobin, but for Sickle-cell Anemia this amino acid is coded for ...
Título 01 Universidade Fernando Pessoa
... • Quick, highly redundant – requires 7-9X coverage for sequencing reads of 500-750bp. This means that for the Human Genome of 3 billion bp, 21-27 billion bases need to be sequence to provide adequate fragment overlap. • Computationally intensive • Troubles with repetitive DNA • Original strategy of ...
... • Quick, highly redundant – requires 7-9X coverage for sequencing reads of 500-750bp. This means that for the Human Genome of 3 billion bp, 21-27 billion bases need to be sequence to provide adequate fragment overlap. • Computationally intensive • Troubles with repetitive DNA • Original strategy of ...
Chapter 16: The Molecular Basis of Inheritance
... 12. What are the functions of primase? DNA polymerase? Ligase? 13. What is the difference between the 5’ and 3’ ends of the DNA molecule? Where are the 5’ and 3’ ends on opposite strands of the double helix? 14. What is the difference between the leading and lagging strand during replication? Why ar ...
... 12. What are the functions of primase? DNA polymerase? Ligase? 13. What is the difference between the 5’ and 3’ ends of the DNA molecule? Where are the 5’ and 3’ ends on opposite strands of the double helix? 14. What is the difference between the leading and lagging strand during replication? Why ar ...
Molecular Genetics Outcome Checklist
... _____ I can explain how, in general, restriction enzymes cut DNA molecules into smaller fragments based on a specific nucleotide sequence, leaving “sticky ends”. _____ I understand the purpose and function of ligases. _____ I can explain how restriction enzymes, ligases, and other DNA technology ca ...
... _____ I can explain how, in general, restriction enzymes cut DNA molecules into smaller fragments based on a specific nucleotide sequence, leaving “sticky ends”. _____ I understand the purpose and function of ligases. _____ I can explain how restriction enzymes, ligases, and other DNA technology ca ...
Let`s Find the Pheromone Gene
... 2. Using pipettor, fill wells with 5uL of Head, Thorax, and Abdomen PCR products as well as the controls and the ladder 3. Molecular Technician puts gel in the buffer-filled box and starts the electrical charge (RUN TO RED! DNA is negative and runs to the positive charge.) Let the gel run for 10 min ...
... 2. Using pipettor, fill wells with 5uL of Head, Thorax, and Abdomen PCR products as well as the controls and the ladder 3. Molecular Technician puts gel in the buffer-filled box and starts the electrical charge (RUN TO RED! DNA is negative and runs to the positive charge.) Let the gel run for 10 min ...
Biotechnology - Hicksville Public Schools / Homepage
... Human Genome Project: In 1990, advances in DNA technology enabled scientists to completely sequence the human genome. A rough draft was complete in 2000. ...
... Human Genome Project: In 1990, advances in DNA technology enabled scientists to completely sequence the human genome. A rough draft was complete in 2000. ...
Chapter 10 Structure and Function of DNA
... Meselson-Stahl experiment – what was the significance? http://highered.mcgrawhill.com/olcweb/cgi/pluginpop.cgi?it=swf::535::535::/sites/dl/free/0072437316/120076/bio22.swf:: Meselson%20and%20Stahl%20Experiment Complementary Base Pairing What is a nucleotide? What is the backbone Where are the 5’ and ...
... Meselson-Stahl experiment – what was the significance? http://highered.mcgrawhill.com/olcweb/cgi/pluginpop.cgi?it=swf::535::535::/sites/dl/free/0072437316/120076/bio22.swf:: Meselson%20and%20Stahl%20Experiment Complementary Base Pairing What is a nucleotide? What is the backbone Where are the 5’ and ...
Journey Into dna
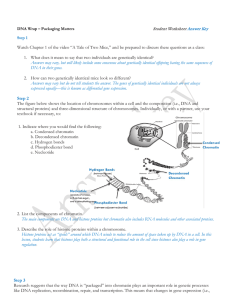
... There is a lot of DNA within the nucleus-about _________feet if you could unravel it and stretch it out. Chromatin scaffold: Chromatin refers to ___________________ that help organize the long DNA molecule. Nucleosome: Double helix: Which two scientists used Rosalind Franklin’s photo 51 to piece tog ...
... There is a lot of DNA within the nucleus-about _________feet if you could unravel it and stretch it out. Chromatin scaffold: Chromatin refers to ___________________ that help organize the long DNA molecule. Nucleosome: Double helix: Which two scientists used Rosalind Franklin’s photo 51 to piece tog ...
Bisulfite sequencing

Bisulphite sequencing (also known as bisulfite sequencing) is the use of bisulphite treatment of DNA to determine its pattern of methylation. DNA methylation was the first discovered epigenetic mark, and remains the most studied. In animals it predominantly involves the addition of a methyl group to the carbon-5 position of cytosine residues of the dinucleotide CpG, and is implicated in repression of transcriptional activity.Treatment of DNA with bisulphite converts cytosine residues to uracil, but leaves 5-methylcytosine residues unaffected. Thus, bisulphite treatment introduces specific changes in the DNA sequence that depend on the methylation status of individual cytosine residues, yielding single- nucleotide resolution information about the methylation status of a segment of DNA. Various analyses can be performed on the altered sequence to retrieve this information. The objective of this analysis is therefore reduced to differentiating between single nucleotide polymorphisms (cytosines and thymidine) resulting from bisulphite conversion (Figure 1).