Genetics - Max Appeal!
... chromosomes is examined under a microscope. This is used for detecting large chromosomal rearrangements or deletions, or extra chromosomes like in Down’s syndrome. The 22q11.2 deletion is too small to be seen by this method so other tests have been developed. The descriptions of these tests are huge ...
... chromosomes is examined under a microscope. This is used for detecting large chromosomal rearrangements or deletions, or extra chromosomes like in Down’s syndrome. The 22q11.2 deletion is too small to be seen by this method so other tests have been developed. The descriptions of these tests are huge ...
Genetics - Max Appeal!
... chromosomes is examined under a microscope. This is used for detecting large chromosomal rearrangements or deletions, or extra chromosomes like in Down’s syndrome. The 22q11.2 deletion is too small to be seen by this method so other tests have been developed. The descriptions of these tests are huge ...
... chromosomes is examined under a microscope. This is used for detecting large chromosomal rearrangements or deletions, or extra chromosomes like in Down’s syndrome. The 22q11.2 deletion is too small to be seen by this method so other tests have been developed. The descriptions of these tests are huge ...
Serological and molecular techniques to detect and identify plant
... has been made possible by the discovery of hybridomas which are fusions between the antibody-producing cells and bone marrow tumour (myeloma) cells. The hybridoma cells multiply in vitro and each culture derived from a single cell produces an epitope-specific antibody. The IgG produced by the one ce ...
... has been made possible by the discovery of hybridomas which are fusions between the antibody-producing cells and bone marrow tumour (myeloma) cells. The hybridoma cells multiply in vitro and each culture derived from a single cell produces an epitope-specific antibody. The IgG produced by the one ce ...
Ch11_Lecture no writing
... DNA can be damaged by radiation, toxic chemicals, and random spontaneous chemical reactions. Excision repair: enzymes constantly scan DNA for mispaired bases, chemically modified bases, and extra bases—unpaired loops. ...
... DNA can be damaged by radiation, toxic chemicals, and random spontaneous chemical reactions. Excision repair: enzymes constantly scan DNA for mispaired bases, chemically modified bases, and extra bases—unpaired loops. ...
Methods to Increase the Percentage of Free Fetal DNA Recovered
... Prenatal diagnosis is useful for managing a pregnancy with an identified fetal abnormality, and may allow for planning and coordinating of care during delivery and the neonatal period.1 A variety of prenatal diagnostic tests are available but each test has limitations. The two most commonly utilized ...
... Prenatal diagnosis is useful for managing a pregnancy with an identified fetal abnormality, and may allow for planning and coordinating of care during delivery and the neonatal period.1 A variety of prenatal diagnostic tests are available but each test has limitations. The two most commonly utilized ...
PCR - UCLA EEB
... 2. Fill out PCR sheet with date, sample numbers, type of extraction, other notes. 3. Multiply out volumes of standard protocol by X (where X= # samples + pos. PCR control + neg. Chelex control + neg.PCR control + 1 extra). 4. Label and number PCR tubes in rack. Set up samples so that they are in a p ...
... 2. Fill out PCR sheet with date, sample numbers, type of extraction, other notes. 3. Multiply out volumes of standard protocol by X (where X= # samples + pos. PCR control + neg. Chelex control + neg.PCR control + 1 extra). 4. Label and number PCR tubes in rack. Set up samples so that they are in a p ...
Sample Chapter
... location (in base pairs) and distance between genes or markers, or unknown DNA or genes. These maps provide information about the physical organization of the DNA; examples are the location of restriction enzyme sites and the order of restriction fragments of chromosomes. An entire genome can be stu ...
... location (in base pairs) and distance between genes or markers, or unknown DNA or genes. These maps provide information about the physical organization of the DNA; examples are the location of restriction enzyme sites and the order of restriction fragments of chromosomes. An entire genome can be stu ...
Molecular
... suspected mutagens. Some of your his- bacteria reverted to a his+ phenotype, but how do you suppose this happened? As you remember, different mutagens affect DNA in different ways. However, even though a specific mutagen will typically cause one type of mutation, this is not necessarily the only mut ...
... suspected mutagens. Some of your his- bacteria reverted to a his+ phenotype, but how do you suppose this happened? As you remember, different mutagens affect DNA in different ways. However, even though a specific mutagen will typically cause one type of mutation, this is not necessarily the only mut ...
3DNA Printer: A Tool for Automated DNA Origami
... source software for developing 3D nanoscale structures automatically by importing a corresponding .obj file which can be obtained from any well known software like Autocad, Maya etc. for any desired 3D structure.Current version of the software designs only Platonic solids. In future, we will release ...
... source software for developing 3D nanoscale structures automatically by importing a corresponding .obj file which can be obtained from any well known software like Autocad, Maya etc. for any desired 3D structure.Current version of the software designs only Platonic solids. In future, we will release ...
Title CHROMOSOMAL ASSIGNMENT OF
... chromosomes l and 2 ( i f they are present) in TAIA and TA3A, as recovered from the fraction shown by an arrow in (A), were analysed s i m i l a r l y . Lanes T and M: total human DNA and total mouse DNA, respectively. ...
... chromosomes l and 2 ( i f they are present) in TAIA and TA3A, as recovered from the fraction shown by an arrow in (A), were analysed s i m i l a r l y . Lanes T and M: total human DNA and total mouse DNA, respectively. ...
Section E
... • Both leading and lagging strand primers are elongated by DNA polymerase III holoenzyme. This complex is a dimer, – One half synthesizes the leading strand; – The other synthesizes the lagging strand; – The two polymerases in a single complex ensures that both strands are synthesized at the same ra ...
... • Both leading and lagging strand primers are elongated by DNA polymerase III holoenzyme. This complex is a dimer, – One half synthesizes the leading strand; – The other synthesizes the lagging strand; – The two polymerases in a single complex ensures that both strands are synthesized at the same ra ...
Breast Cancer: FISH for ERBB2 (HER2)
... monoclonal antibody Herceptin is contemplated. The results should be interpreted in context with other genetic, pathological, and clinical information. ...
... monoclonal antibody Herceptin is contemplated. The results should be interpreted in context with other genetic, pathological, and clinical information. ...
standard set 5 - EDHSGreenSea.net
... 1. In the 1930s the favored hypothesis suggested that the genetic material (the chemical substance that carried hereditary information) most probably was protein. Nucleic acids were considered too simple to provide much information and were thought to be structural molecules onto which the informati ...
... 1. In the 1930s the favored hypothesis suggested that the genetic material (the chemical substance that carried hereditary information) most probably was protein. Nucleic acids were considered too simple to provide much information and were thought to be structural molecules onto which the informati ...
Chapter 16 The Molecular Basis of Inheritance
... The final error rate is only one per ten billion nucleotides. DNA molecules are constantly subject to potentially harmful chemical and physical agents. Reactive chemicals, radioactive emissions, X-rays, and ultraviolet light can change nucleotides in ways that can affect encoded genetic inform ...
... The final error rate is only one per ten billion nucleotides. DNA molecules are constantly subject to potentially harmful chemical and physical agents. Reactive chemicals, radioactive emissions, X-rays, and ultraviolet light can change nucleotides in ways that can affect encoded genetic inform ...
The Molecular Basis of Inheritance
... The final error rate is only one per ten billion nucleotides. DNA molecules are constantly subject to potentially harmful chemical and physical agents. Reactive chemicals, radioactive emissions, X-rays, and ultraviolet light can change nucleotides in ways that can affect encoded genetic inform ...
... The final error rate is only one per ten billion nucleotides. DNA molecules are constantly subject to potentially harmful chemical and physical agents. Reactive chemicals, radioactive emissions, X-rays, and ultraviolet light can change nucleotides in ways that can affect encoded genetic inform ...
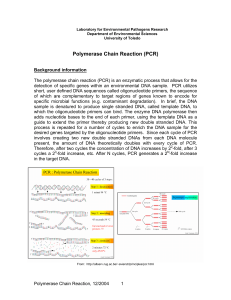
Polymerase Chain Reaction (PCR) - Department of Environmental
... Template DNA (target gene) This is the DNA/gene that you wish to amplify. The default concentration of DNA used in our laboratory is 1 ng µl-1 of PCR reaction. However, this concentration can vary by a few orders of magnitude depending on the target gene concentration and source of DNA. Higher amoun ...
... Template DNA (target gene) This is the DNA/gene that you wish to amplify. The default concentration of DNA used in our laboratory is 1 ng µl-1 of PCR reaction. However, this concentration can vary by a few orders of magnitude depending on the target gene concentration and source of DNA. Higher amoun ...
CHAPTER 16 THE MOLECULE BASIS OF INHERITANCE
... In their experiments, they labeled the nucleotides of the old strands with a heavy isotope of nitrogen (15N), while any new nucleotides were indicated by a lighter isotope (14N). Replicated strands could be separated by density in a centrifuge. Each model—the semiconservative model, the conser ...
... In their experiments, they labeled the nucleotides of the old strands with a heavy isotope of nitrogen (15N), while any new nucleotides were indicated by a lighter isotope (14N). Replicated strands could be separated by density in a centrifuge. Each model—the semiconservative model, the conser ...
DNA Replication
... 2. A free 3'OH group is required for replication, but when the two chains separate no group of that nature exists. RNA primers are synthesized, and the free 3'OH of the primer is used to begin replication. 3. The replication fork moves in one direction, but DNA replication only goes in the 5' to 3' ...
... 2. A free 3'OH group is required for replication, but when the two chains separate no group of that nature exists. RNA primers are synthesized, and the free 3'OH of the primer is used to begin replication. 3. The replication fork moves in one direction, but DNA replication only goes in the 5' to 3' ...
Comparative genomic hybridization

Comparative genomic hybridization is a molecular cytogenetic method for analysing copy number variations (CNVs) relative to ploidy level in the DNA of a test sample compared to a reference sample, without the need for culturing cells. The aim of this technique is to quickly and efficiently compare two genomic DNA samples arising from two sources, which are most often closely related, because it is suspected that they contain differences in terms of either gains or losses of either whole chromosomes or subchromosomal regions (a portion of a whole chromosome). This technique was originally developed for the evaluation of the differences between the chromosomal complements of solid tumor and normal tissue, and has an improved resoIution of 5-10 megabases compared to the more traditional cytogenetic analysis techniques of giemsa banding and fluorescence in situ hybridization (FISH) which are limited by the resolution of the microscope utilized.This is achieved through the use of competitive fluorescence in situ hybridization. In short, this involves the isolation of DNA from the two sources to be compared, most commonly a test and reference source, independent labelling of each DNA sample with a different fluorophores (fluorescent molecules) of different colours (usually red and green), denaturation of the DNA so that it is single stranded, and the hybridization of the two resultant samples in a 1:1 ratio to a normal metaphase spread of chromosomes, to which the labelled DNA samples will bind at their locus of origin. Using a fluorescence microscope and computer software, the differentially coloured fluorescent signals are then compared along the length of each chromosome for identification of chromosomal differences between the two sources. A higher intensity of the test sample colour in a specific region of a chromosome indicates the gain of material of that region in the corresponding source sample, while a higher intensity of the reference sample colour indicates the loss of material in the test sample in that specific region. A neutral colour (yellow when the fluorophore labels are red and green) indicates no difference between the two samples in that location.CGH is only able to detect unbalanced chromosomal abnormalities. This is because balanced chromosomal abnormalities such as reciprocal translocations, inversions or ring chromosomes do not affect copy number, which is what is detected by CGH technologies. CGH does, however, allow for the exploration of all 46 human chromosomes in single test and the discovery of deletions and duplications, even on the microscopic scale which may lead to the identification of candidate genes to be further explored by other cytological techniques.Through the use of DNA microarrays in conjunction with CGH techniques, the more specific form of array CGH (aCGH) has been developed, allowing for a locus-by-locus measure of CNV with increased resolution as low as 100 kilobases. This improved technique allows for the aetiology of known and unknown conditions to be discovered.