DNA Structure and Replication
... DNA chains can be very long ! E. coli chromosome: 4.6x106 base pairs: 4.6x 106 x .34 nm = 1.5x106 nm = 1.5 mm ! Human DNA: 6x10-12 g/cell x 1/660 mol bp/g x 6.023x1023 bp/mol bp x 0.34x10-9 m/bp = 1.9 m ! Bacterial, viral DNA “chromosomes” are circles ! DNA in human chromosomes (and DNA of all euka ...
... DNA chains can be very long ! E. coli chromosome: 4.6x106 base pairs: 4.6x 106 x .34 nm = 1.5x106 nm = 1.5 mm ! Human DNA: 6x10-12 g/cell x 1/660 mol bp/g x 6.023x1023 bp/mol bp x 0.34x10-9 m/bp = 1.9 m ! Bacterial, viral DNA “chromosomes” are circles ! DNA in human chromosomes (and DNA of all euka ...
DNA: Structure and Function
... 1940’s-1960’s that indicated DNA was the Molecule of Heredity • Griffith & Avery—DNA transformed nonvirulent bacteria to virulent bacteria • Hershey & Chase—DNA from viruses is injected to host bacteria cells, cells become ...
... 1940’s-1960’s that indicated DNA was the Molecule of Heredity • Griffith & Avery—DNA transformed nonvirulent bacteria to virulent bacteria • Hershey & Chase—DNA from viruses is injected to host bacteria cells, cells become ...
Chapter 15 – Recombinant DNA and Genetic Engineering
... fragments through a gel – DNA is negative – Size of fragment determines how far it migrates • The fewer tandem repeats the farther it travels • Differences in homologous DNA sequences resulting in fragments of different lengths are restriction fragment length polymorphisms (RFLP’s) ...
... fragments through a gel – DNA is negative – Size of fragment determines how far it migrates • The fewer tandem repeats the farther it travels • Differences in homologous DNA sequences resulting in fragments of different lengths are restriction fragment length polymorphisms (RFLP’s) ...
11/01 Molecular genetic analysis and biotechnology
... Cloning Genes • Gene cloning: amplifying a specific piece of DNA via a bacteria cell • Cloning vector: a replicating DNA molecule attached with a foreign DNA fragment to be introduced into a cell – Has features that make it easier to insert DNA and select for presence of vector in cell. • Origin o ...
... Cloning Genes • Gene cloning: amplifying a specific piece of DNA via a bacteria cell • Cloning vector: a replicating DNA molecule attached with a foreign DNA fragment to be introduced into a cell – Has features that make it easier to insert DNA and select for presence of vector in cell. • Origin o ...
Lecture 2: Biology Review II
... short unique fragment of DNA. Definition: Expressed sequence tags (EST) are subsets of STSs from cDNA clones. Represent transcribed genes (e.g. usually proteins). ...
... short unique fragment of DNA. Definition: Expressed sequence tags (EST) are subsets of STSs from cDNA clones. Represent transcribed genes (e.g. usually proteins). ...
Molecluar Genetics Key
... An anti-sense strand of DNA has bases ATC GAT CCG. Which is the correct sequence for bases on tRNA anticodons coded from this DNA? (A) (B) (C) (D) ...
... An anti-sense strand of DNA has bases ATC GAT CCG. Which is the correct sequence for bases on tRNA anticodons coded from this DNA? (A) (B) (C) (D) ...
Comparative Genome Organization in plants: From Sequence and Markers to... and Chromosomes Summary
... variation within the motifs should be known in order to ascertain the function of the repeat elements in the genome. Some of the sequence repeats have been highly conserved from one species to another like the rDNA genes, but some repeats are highly variable even between accessions of a species. The ...
... variation within the motifs should be known in order to ascertain the function of the repeat elements in the genome. Some of the sequence repeats have been highly conserved from one species to another like the rDNA genes, but some repeats are highly variable even between accessions of a species. The ...
DNA Extraction
... strategy ensures the integrity of the code, for the proteins that result from the nucleotide sequence are vital to the cell. Every cell that comprises a living organism contains the complete genetic blueprint of that organism, what enables the specialization of a particular cell in a particular area ...
... strategy ensures the integrity of the code, for the proteins that result from the nucleotide sequence are vital to the cell. Every cell that comprises a living organism contains the complete genetic blueprint of that organism, what enables the specialization of a particular cell in a particular area ...
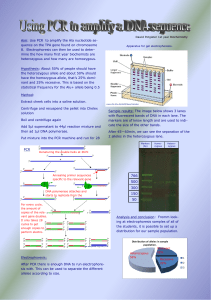
PCR
... Hypothesis: About 50% of people should have the heterozygous allele and about 50% should have the homozygous allele, that’s 25% dominant and 25% recessive. This is based on the statistical frequency for the Alu+ allele being 0.5 Method: Extract cheek cells into a saline solution. ...
... Hypothesis: About 50% of people should have the heterozygous allele and about 50% should have the homozygous allele, that’s 25% dominant and 25% recessive. This is based on the statistical frequency for the Alu+ allele being 0.5 Method: Extract cheek cells into a saline solution. ...
Course Outline - Pima Community College
... Upon completion of the course, the student will be able to do the following: 1. Implement the scientific method to answer specific questions. 2. Calibrate and use a variety of common and basic types of lab equipment. 3. Read and follow an instruction manual for lab equipment and lab procedures. 4. C ...
... Upon completion of the course, the student will be able to do the following: 1. Implement the scientific method to answer specific questions. 2. Calibrate and use a variety of common and basic types of lab equipment. 3. Read and follow an instruction manual for lab equipment and lab procedures. 4. C ...
appendix h: detection and significance of genetic abnormalities
... Studies in the chemically exposed show that this is quite a reproducible method of looking at DNA damage. However, other factors known to influence results are increasing age, gender and smoking as well as the degree of exposure to chemicals, plasma B12 and plasma homocysteine levels. There is also ...
... Studies in the chemically exposed show that this is quite a reproducible method of looking at DNA damage. However, other factors known to influence results are increasing age, gender and smoking as well as the degree of exposure to chemicals, plasma B12 and plasma homocysteine levels. There is also ...
forensic_biology
... sequencing. Like any DNA fragment, SSRs can be detected by specific dyes or by radiolabelling using gel electrophoresis. The advantage of using SSRs as molecular markers is the extent of polymorphism shown, which enables the detection of differences at multiple loci between strains [3].Coupled with ...
... sequencing. Like any DNA fragment, SSRs can be detected by specific dyes or by radiolabelling using gel electrophoresis. The advantage of using SSRs as molecular markers is the extent of polymorphism shown, which enables the detection of differences at multiple loci between strains [3].Coupled with ...
Biotechnology Pre/PostTest Key (w/citations)
... _____4) The rate of migration of DNA within an agarose gel during electrophoresis is primarily based on what factor?? A. The size of the DNA fragments. B. The size of the wells in the gel. C. The volume of the DNA sample loaded D. The number of DNA fragments. 2012 FL Holt McDougal Biology ...
... _____4) The rate of migration of DNA within an agarose gel during electrophoresis is primarily based on what factor?? A. The size of the DNA fragments. B. The size of the wells in the gel. C. The volume of the DNA sample loaded D. The number of DNA fragments. 2012 FL Holt McDougal Biology ...
Comparative genomic hybridization

Comparative genomic hybridization is a molecular cytogenetic method for analysing copy number variations (CNVs) relative to ploidy level in the DNA of a test sample compared to a reference sample, without the need for culturing cells. The aim of this technique is to quickly and efficiently compare two genomic DNA samples arising from two sources, which are most often closely related, because it is suspected that they contain differences in terms of either gains or losses of either whole chromosomes or subchromosomal regions (a portion of a whole chromosome). This technique was originally developed for the evaluation of the differences between the chromosomal complements of solid tumor and normal tissue, and has an improved resoIution of 5-10 megabases compared to the more traditional cytogenetic analysis techniques of giemsa banding and fluorescence in situ hybridization (FISH) which are limited by the resolution of the microscope utilized.This is achieved through the use of competitive fluorescence in situ hybridization. In short, this involves the isolation of DNA from the two sources to be compared, most commonly a test and reference source, independent labelling of each DNA sample with a different fluorophores (fluorescent molecules) of different colours (usually red and green), denaturation of the DNA so that it is single stranded, and the hybridization of the two resultant samples in a 1:1 ratio to a normal metaphase spread of chromosomes, to which the labelled DNA samples will bind at their locus of origin. Using a fluorescence microscope and computer software, the differentially coloured fluorescent signals are then compared along the length of each chromosome for identification of chromosomal differences between the two sources. A higher intensity of the test sample colour in a specific region of a chromosome indicates the gain of material of that region in the corresponding source sample, while a higher intensity of the reference sample colour indicates the loss of material in the test sample in that specific region. A neutral colour (yellow when the fluorophore labels are red and green) indicates no difference between the two samples in that location.CGH is only able to detect unbalanced chromosomal abnormalities. This is because balanced chromosomal abnormalities such as reciprocal translocations, inversions or ring chromosomes do not affect copy number, which is what is detected by CGH technologies. CGH does, however, allow for the exploration of all 46 human chromosomes in single test and the discovery of deletions and duplications, even on the microscopic scale which may lead to the identification of candidate genes to be further explored by other cytological techniques.Through the use of DNA microarrays in conjunction with CGH techniques, the more specific form of array CGH (aCGH) has been developed, allowing for a locus-by-locus measure of CNV with increased resolution as low as 100 kilobases. This improved technique allows for the aetiology of known and unknown conditions to be discovered.