Introduction to Gel Electrophorsis
... hydrolysis : H2O H+ and OH• The anode (+ /red) pole becomes alkaline because OH- will accumulate at this pole • The cathode (-/black) pole becomes acidic because H+ will accumulate at this pole ...
... hydrolysis : H2O H+ and OH• The anode (+ /red) pole becomes alkaline because OH- will accumulate at this pole • The cathode (-/black) pole becomes acidic because H+ will accumulate at this pole ...
Pursuing DNA Catalysts for Protein Modification
... groups when presented on a protein surface. Also, smallmolecule catalysts may not tolerate the aqueous conditions required to function with most protein substrates. Evolved protein enzymes have achieved many successes, but if nature has not provided a reasonably good enzymatic starting point for a d ...
... groups when presented on a protein surface. Also, smallmolecule catalysts may not tolerate the aqueous conditions required to function with most protein substrates. Evolved protein enzymes have achieved many successes, but if nature has not provided a reasonably good enzymatic starting point for a d ...
Industrial Biotechnology
... • Some enzymes are produced only when their substrate is available in the medium. (inducible enzymes). • Analogues of the substrate may act as the inducer. • When an inducer is present in the medium a number of different inducible enzymes may sometimes be synthesized by the organism. • The pathway f ...
... • Some enzymes are produced only when their substrate is available in the medium. (inducible enzymes). • Analogues of the substrate may act as the inducer. • When an inducer is present in the medium a number of different inducible enzymes may sometimes be synthesized by the organism. • The pathway f ...
Enzymopathy as a result of Polymorphism
... Disabled Enzymes Enzymes may also be disabled as a result of polymorphisms. When polymorphic amino acid sequences are present in the enzymes, the enzymes are inadequate for transfer of the biochemical unit to a product. In essence, the enzyme is disabled and therefore cannot facilitate product produ ...
... Disabled Enzymes Enzymes may also be disabled as a result of polymorphisms. When polymorphic amino acid sequences are present in the enzymes, the enzymes are inadequate for transfer of the biochemical unit to a product. In essence, the enzyme is disabled and therefore cannot facilitate product produ ...
to an allosteric site
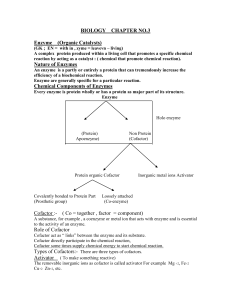
... enzyme's three-dimensional shape. Substrate = The substance an enzyme acts on and makes more reactive. • An enzyme binds to its substrate and catalyzes its conversion to product. The enzyme is released in original form. Substrate + enzyme enzyme-substrate complex product + enzyme • The substrate bin ...
... enzyme's three-dimensional shape. Substrate = The substance an enzyme acts on and makes more reactive. • An enzyme binds to its substrate and catalyzes its conversion to product. The enzyme is released in original form. Substrate + enzyme enzyme-substrate complex product + enzyme • The substrate bin ...
Chemistry 20 Chapters 15 Enzymes
... than the active site but that eventually affects the active site. This type of interaction is called allosterism, and any enzyme regulated by this mechanism is called an allosteric enzyme. If a substance binds noncovalently and reversibly to a site other the active site, it may affect the enzyme in ...
... than the active site but that eventually affects the active site. This type of interaction is called allosterism, and any enzyme regulated by this mechanism is called an allosteric enzyme. If a substance binds noncovalently and reversibly to a site other the active site, it may affect the enzyme in ...
AZT resistance of simian foamy virus reverse transcriptase is based
... C-terminus. To avoid degradation of the PR–RT by autocatalytic activity of the PR, a mutant enzyme was constructed harboring an active site mutation in the PR region (D24A), which leads to an inactive PR. Thus the AZT-resistant mutants also contain the D24A mutation in the PR. The AZT-resistance mut ...
... C-terminus. To avoid degradation of the PR–RT by autocatalytic activity of the PR, a mutant enzyme was constructed harboring an active site mutation in the PR region (D24A), which leads to an inactive PR. Thus the AZT-resistant mutants also contain the D24A mutation in the PR. The AZT-resistance mut ...
active site - Blue Valley Schools
... The reactant that an enzyme acts on is called the enzyme’s substrate. The enzyme binds to its substrate, forming an enzyme-substrate complex. The active site is the region on the enzyme where the substrate binds. Induced fit of a substrate brings chemical groups of the active site into positi ...
... The reactant that an enzyme acts on is called the enzyme’s substrate. The enzyme binds to its substrate, forming an enzyme-substrate complex. The active site is the region on the enzyme where the substrate binds. Induced fit of a substrate brings chemical groups of the active site into positi ...
Flavin adenine dinucleotide as a chromophore of the Xenopus (6
... the bases to their native forrn (7). In this reaction, the near-UV/blue light photon is used to excite FADI-I- and flavin in the excited state then donates an electron to the CPD and thus FAD is essential for the reaction. The CPD photolyase gene has been isolated from 13 organisms and, on the basis ...
... the bases to their native forrn (7). In this reaction, the near-UV/blue light photon is used to excite FADI-I- and flavin in the excited state then donates an electron to the CPD and thus FAD is essential for the reaction. The CPD photolyase gene has been isolated from 13 organisms and, on the basis ...
Biochemical and functional characterization of Plasmodium
... DNA polymerase δ is an essential enzyme required for chromosomal DNA replication and repair, and therefore may be a potential target for anti-malarial drug development. However, little is known of the characteristics and function of this P. falciparum enzyme. Methods: The coding sequences of DNA po ...
... DNA polymerase δ is an essential enzyme required for chromosomal DNA replication and repair, and therefore may be a potential target for anti-malarial drug development. However, little is known of the characteristics and function of this P. falciparum enzyme. Methods: The coding sequences of DNA po ...
Evolution by leaps: gene duplication in bacteria | SpringerLink
... Background: Sequence related families of genes and proteins are common in bacterial genomes. In Escherichia coli they constitute over half of the genome. The presence of families and superfamilies of proteins suggest a history of gene duplication and divergence during evolution. Genome encoded prote ...
... Background: Sequence related families of genes and proteins are common in bacterial genomes. In Escherichia coli they constitute over half of the genome. The presence of families and superfamilies of proteins suggest a history of gene duplication and divergence during evolution. Genome encoded prote ...
Chapter 6 "Mechanisms of Enzymes" Reading Assignment: pp. 158
... transition state structure, chemicals known as transition-state analogs, which resemble the structure of the transition state, have been shown to bind to enzymes with higher affinity than substrates. Due to tight binding, many of these molecules are good inhibitors of enzymes. For example, the antib ...
... transition state structure, chemicals known as transition-state analogs, which resemble the structure of the transition state, have been shown to bind to enzymes with higher affinity than substrates. Due to tight binding, many of these molecules are good inhibitors of enzymes. For example, the antib ...
pdf-download
... racemic resolution of amino acids like methionine to the desired enantiopure L-compounds. Another class of enzymes that can be found regularly in commercialised processes are the so-called oxidoreductases, enzymes that perform oxidation or reduction reactions. A well-known example is Eli Lilly’s who ...
... racemic resolution of amino acids like methionine to the desired enantiopure L-compounds. Another class of enzymes that can be found regularly in commercialised processes are the so-called oxidoreductases, enzymes that perform oxidation or reduction reactions. A well-known example is Eli Lilly’s who ...