TCSS Biology Unit 2 – Genetics Information
... Bioethics: Your Genes, Your Choice Comprehension Questions Group activity and comprehension questions that gives students scenarios in which they have to decide what they would do with medical information. Exploring Gene Therapy WebQuest - Student comprehension questions about the basic of gene ther ...
... Bioethics: Your Genes, Your Choice Comprehension Questions Group activity and comprehension questions that gives students scenarios in which they have to decide what they would do with medical information. Exploring Gene Therapy WebQuest - Student comprehension questions about the basic of gene ther ...
Mismatch repair
... nick, removing a portion of the damaged strand (with its 5’3’ exonuclease activity) and replacing it with undamaged DNA. (d) The nick remaining after DNA polymerase I has dissociated is sealed by DNA ligase. ...
... nick, removing a portion of the damaged strand (with its 5’3’ exonuclease activity) and replacing it with undamaged DNA. (d) The nick remaining after DNA polymerase I has dissociated is sealed by DNA ligase. ...
I. DNA, Chromosomes, Chromatin, and Genes II. DNA
... DNA & Protein Synthesis Review Worksheet 1) A _________________________ is a segment of DNA that codes for a protein. 2) __________________________________________ is uncoiled DNA. 3) __________________________________________ is coiled DNA. 4) _________________________________________ is the enzym ...
... DNA & Protein Synthesis Review Worksheet 1) A _________________________ is a segment of DNA that codes for a protein. 2) __________________________________________ is uncoiled DNA. 3) __________________________________________ is coiled DNA. 4) _________________________________________ is the enzym ...
13059_2010_2366_MOESM1_ESM
... scaffolds that were comprised of 1,747 contigs with an average depth of coverage of ~8X. The N50 contig length is 124 kb and the N50 scaffold length is 773,464 bp. There are 772 scaffold gaps comprising 2.1Mb of N’s in the scaffolds which represent sequencing gaps. The GC percentage of the genome is ...
... scaffolds that were comprised of 1,747 contigs with an average depth of coverage of ~8X. The N50 contig length is 124 kb and the N50 scaffold length is 773,464 bp. There are 772 scaffold gaps comprising 2.1Mb of N’s in the scaffolds which represent sequencing gaps. The GC percentage of the genome is ...
Biotechnology Laboratory
... 1) Data Analysis and Critical Thinking. Special emphasis will be placed on the ability of graduate students to understand and interpret data and think analytically and critically about information necessary to understand and perform lab experiments. Graduate students are further expected to develop ...
... 1) Data Analysis and Critical Thinking. Special emphasis will be placed on the ability of graduate students to understand and interpret data and think analytically and critically about information necessary to understand and perform lab experiments. Graduate students are further expected to develop ...
Document
... • DNA polymerases are key enzymes in replication • once the two strands have separated at the replication fork, the nucleotides must be lined up in proper order for DNA synthesis • in the absence of DNA polymerase, alignment is slow • DNA polymerase provides the speed and specificity of alignment • ...
... • DNA polymerases are key enzymes in replication • once the two strands have separated at the replication fork, the nucleotides must be lined up in proper order for DNA synthesis • in the absence of DNA polymerase, alignment is slow • DNA polymerase provides the speed and specificity of alignment • ...
DNA purification and isolation of genomic DNA from bacterial
... overall versatility and consistency. This technique exploits the difference in denaturation and renaturation characteristics of covalently closed circular plasmid DNA and chromosomal DNA fragments. Under alkaline conditions (at pH 11), both plasmid and chromosomal DNA are efficiently denatured. Rapi ...
... overall versatility and consistency. This technique exploits the difference in denaturation and renaturation characteristics of covalently closed circular plasmid DNA and chromosomal DNA fragments. Under alkaline conditions (at pH 11), both plasmid and chromosomal DNA are efficiently denatured. Rapi ...
Transduction
... There is no meiosis in bacteria so special techniques have been worked out for manipulating genes in bacteria so that mapping experiments, strain construction, and complementation tests can be done. First, we need a way of getting chromosomal DNA from one cell into another. There are several ways to ...
... There is no meiosis in bacteria so special techniques have been worked out for manipulating genes in bacteria so that mapping experiments, strain construction, and complementation tests can be done. First, we need a way of getting chromosomal DNA from one cell into another. There are several ways to ...
Trans-HHS Workshop: Diet, DNA Methylation
... enzyme was associated with elevated plasma homocysteine levels, which is to be expected because the conversion of homocysteine to methionine is impaired (31). Not only did the mild hyperhomocystinemia appear as an indicator of altered one-carbon metabolism, but the higher levels of this sulfur-conta ...
... enzyme was associated with elevated plasma homocysteine levels, which is to be expected because the conversion of homocysteine to methionine is impaired (31). Not only did the mild hyperhomocystinemia appear as an indicator of altered one-carbon metabolism, but the higher levels of this sulfur-conta ...
2016‐12‐15 1
... What is the difference between a chromosome and a chromatid? A replicated chromosome is made up of two chromatids which are joined by the centromere. The chromatids separate from each other during mitosis and is dispersed as chromatin during mitosis. What are chromosome homologs? One chromosome of ...
... What is the difference between a chromosome and a chromatid? A replicated chromosome is made up of two chromatids which are joined by the centromere. The chromatids separate from each other during mitosis and is dispersed as chromatin during mitosis. What are chromosome homologs? One chromosome of ...
GMM Risk Assessment - Queen`s University Belfast
... /deleted/complemented Gene(s) Genes should be identified so that reviewers have a general idea of their function (a 3 letter name may be insufficient) Where gene function is not known please give details of any known homologues. Generic examples may be sufficient. 2.5 Most Hazardous GMM Considering ...
... /deleted/complemented Gene(s) Genes should be identified so that reviewers have a general idea of their function (a 3 letter name may be insufficient) Where gene function is not known please give details of any known homologues. Generic examples may be sufficient. 2.5 Most Hazardous GMM Considering ...
Chapter 20
... • A clone carrying the gene of interest can be identified with a nucleic acid probe having a sequence complementary to the gene – Since we know the sequence of the fruit fly gene we can make a short primer and use this to “probe” our library • This process is called nucleic acid ...
... • A clone carrying the gene of interest can be identified with a nucleic acid probe having a sequence complementary to the gene – Since we know the sequence of the fruit fly gene we can make a short primer and use this to “probe” our library • This process is called nucleic acid ...
2006
... substitutions were most often silent (third codon) indicating that most were not generated during the amplification and cloning procedure, which would be blind to codon position. The amino acid substitutions are not evenly distributed across the entire exon, but are more common in the high-glycine ( ...
... substitutions were most often silent (third codon) indicating that most were not generated during the amplification and cloning procedure, which would be blind to codon position. The amino acid substitutions are not evenly distributed across the entire exon, but are more common in the high-glycine ( ...
Gene targeting: vector design and construction
... • Which chromosome? Three copies of chromosome #2; single copy of chromosome Z in DT40, which was derived from female chicken. • Analyze/map genomic structure (exon-intron) of your gene of interest. – don’t trust the database too much! There could be mistakes, differences (polymorphisms) and SNPs. ...
... • Which chromosome? Three copies of chromosome #2; single copy of chromosome Z in DT40, which was derived from female chicken. • Analyze/map genomic structure (exon-intron) of your gene of interest. – don’t trust the database too much! There could be mistakes, differences (polymorphisms) and SNPs. ...
Question paper - Unit F215 - Control, genomes and
... QUESTION 3 STARTS ON PAGE 10 PLEASE DO NOT WRITE ON THIS PAGE ...
... QUESTION 3 STARTS ON PAGE 10 PLEASE DO NOT WRITE ON THIS PAGE ...
APOC3 rs2854116 single nucleotide polymorphism
... allelic discrimination assays [15] but this method is in general costly. The principal method used to determine APOC3 genotypes was Polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP) because this method can be applied in laboratory with uncomplicated and low-cost equipment ...
... allelic discrimination assays [15] but this method is in general costly. The principal method used to determine APOC3 genotypes was Polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP) because this method can be applied in laboratory with uncomplicated and low-cost equipment ...
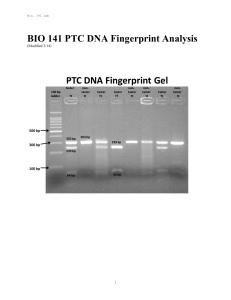
BIO 141 PTC DNA Fingerprint Analysis
... known individual can be matched to a sample from an unidentified individual. Several methods to detect DNA differences exist in DNA fingerprint analysis. Basically, scientists isolate DNA from individuals, measure the size of certain variable fragments of DNA, and compare these DNA fragments to each ...
... known individual can be matched to a sample from an unidentified individual. Several methods to detect DNA differences exist in DNA fingerprint analysis. Basically, scientists isolate DNA from individuals, measure the size of certain variable fragments of DNA, and compare these DNA fragments to each ...
Characterization of the IEll0 Gene of Herpes Simplex Virus Type 1
... As shown in Fig. 1, IE gene 1 is located in the RL element of the HSV-1 genome. We have determined the complete sequence of RL, together with adjacent regions of UL: the whole sequence will be presented elsewhere (L. J. Perry & D. J. McGeoch, unpublished). Residue numbering in this paper is based on ...
... As shown in Fig. 1, IE gene 1 is located in the RL element of the HSV-1 genome. We have determined the complete sequence of RL, together with adjacent regions of UL: the whole sequence will be presented elsewhere (L. J. Perry & D. J. McGeoch, unpublished). Residue numbering in this paper is based on ...
Genomic library

A genomic library is a collection of the total genomic DNA from a single organism. The DNA is stored in a population of identical vectors, each containing a different insert of DNA. In order to construct a genomic library, the organism's DNA is extracted from cells and then digested with a restriction enzyme to cut the DNA into fragments of a specific size. The fragments are then inserted into the vector using DNA ligase. Next, the vector DNA can be taken up by a host organism - commonly a population of Escherichia coli or yeast - with each cell containing only one vector molecule. Using a host cell to carry the vector allows for easy amplification and retrieval of specific clones from the library for analysis.There are several kinds of vectors available with various insert capacities. Generally, libraries made from organisms with larger genomes require vectors featuring larger inserts, thereby fewer vector molecules are needed to make the library. Researchers can choose a vector also considering the ideal insert size to find a desired number of clones necessary for full genome coverage.Genomic libraries are commonly used for sequencing applications. They have played an important role in the whole genome sequencing of several organisms, including the human genome and several model organisms.