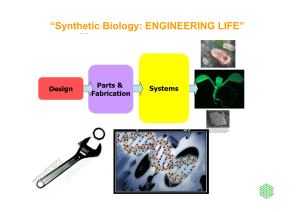
Nomenclature of Transposable Elements in Prokaryotes
... be designated according to the rules de- strains carrying known elements at unveloped for plasmid nomenclature (Novick et known sites are most conveniently treated al., 1976). In particular, each plasmid line as described above for insertions into a parderived from an independent insertion event tic ...
... be designated according to the rules de- strains carrying known elements at unveloped for plasmid nomenclature (Novick et known sites are most conveniently treated al., 1976). In particular, each plasmid line as described above for insertions into a parderived from an independent insertion event tic ...
Tutorial: chloroplast genomes - DOGMA: Annotation of Chloroplast
... strand) as well as within the sequence. To choose a different start codon than the end of the BLAST hit, click on the link and it will change the end of the gene. The start codon for psbA is correct, so we don’t need to change it. You may also change the start and end of the gene manually by typing ...
... strand) as well as within the sequence. To choose a different start codon than the end of the BLAST hit, click on the link and it will change the end of the gene. The start codon for psbA is correct, so we don’t need to change it. You may also change the start and end of the gene manually by typing ...
Mutation
... Experimental test of Lamarck’s “inheritance of acquired traits” Salvador Luria and Max Delbrück (1943) The two possibilities tested by the Luria–Delbrück experiment… (A) If mutations are induced by the media, roughly the same number of mutants are expected to appear on each plate (LAMARCK) (B) If m ...
... Experimental test of Lamarck’s “inheritance of acquired traits” Salvador Luria and Max Delbrück (1943) The two possibilities tested by the Luria–Delbrück experiment… (A) If mutations are induced by the media, roughly the same number of mutants are expected to appear on each plate (LAMARCK) (B) If m ...
The glpP and glpF genes of the glycerol regulon in
... the transcriptional start point for glpFK were identified. In the 5' untranslated leader sequence (UTL) of glpFK mRNA a conserved inverted repeat is found. The repeat is believed to be involved in the control of expression of glpFK by termination/antitermination of transcription, a control mechanism ...
... the transcriptional start point for glpFK were identified. In the 5' untranslated leader sequence (UTL) of glpFK mRNA a conserved inverted repeat is found. The repeat is believed to be involved in the control of expression of glpFK by termination/antitermination of transcription, a control mechanism ...
No Slide Title
... against all reads in the database. Annotation file Similar to GeneSNP-VISTA annotation file.
Recombination points (Optional)
This file must be a tab-delimited file with four fields on each line, in the format:
Sample input files are availab ...
... against all reads in the database. Annotation file Similar to GeneSNP-VISTA annotation file.
Partnership
... The outcome of this work package is knowledge of genes that control recombination frequencies, which, in addition provides molecular markers that allow „breeding‟ with alleles that have either an increase of recombination frequency or suppression. 3) Targeted mutagenesis during meiosis. Methodology ...
... The outcome of this work package is knowledge of genes that control recombination frequencies, which, in addition provides molecular markers that allow „breeding‟ with alleles that have either an increase of recombination frequency or suppression. 3) Targeted mutagenesis during meiosis. Methodology ...
Finding the wheat homologues of genes from model organisms
... The orthologues link under the Plant Compara tool set is an alternative route to find homologous wheat gene for your GOI. It presents a list of all the homologues of any particular gene in other species. The advantage of using the orthologue link is that it reduces the view complexity that could be ...
... The orthologues link under the Plant Compara tool set is an alternative route to find homologous wheat gene for your GOI. It presents a list of all the homologues of any particular gene in other species. The advantage of using the orthologue link is that it reduces the view complexity that could be ...
Computational Biology
... Multiple Genome Rearrangement Problem Find a phylogenetic tree describing the most „plausible“ rearrangement scenario for multiple species. The genomic distance in the case of genome rearrangement is defined in terms of (1) reversals, (2) translocations, (3) fusions, and (4) fissions which are the ...
... Multiple Genome Rearrangement Problem Find a phylogenetic tree describing the most „plausible“ rearrangement scenario for multiple species. The genomic distance in the case of genome rearrangement is defined in terms of (1) reversals, (2) translocations, (3) fusions, and (4) fissions which are the ...
Section D - Prokaryotic and Eukaryotic Chromosome Structure
... • Many applications of genetic engineering require vectors for the expression of genes in diverse eukaryotic species. ...
... • Many applications of genetic engineering require vectors for the expression of genes in diverse eukaryotic species. ...
Folie 1 - ERA-NET PathoGenoMics
... • Type II IFN (IFN-g) activates macrophages and enhances immunity to predominantly nonviral pathogens, particularly when intracellular. • Type I IFN (>10 genes) mediate antiviral innate immunity. It is unclear why their synthesis is an obligatory response to many or even most nonviral pathogens. • T ...
... • Type II IFN (IFN-g) activates macrophages and enhances immunity to predominantly nonviral pathogens, particularly when intracellular. • Type I IFN (>10 genes) mediate antiviral innate immunity. It is unclear why their synthesis is an obligatory response to many or even most nonviral pathogens. • T ...
Optimizing Restriction Site Placement for Synthetic
... Each occurrence of a pattern within a given DNA target sequence is called a restriction enzyme recognition site or restriction site. Unique restriction sites within a given target are particularly prized, as they cut the sequence unambiguously in exactly one place. Many techniques for manipulating ...
... Each occurrence of a pattern within a given DNA target sequence is called a restriction enzyme recognition site or restriction site. Unique restriction sites within a given target are particularly prized, as they cut the sequence unambiguously in exactly one place. Many techniques for manipulating ...
What does the apicoplast do?
... • Apicomplexa appear to have three genomes • Apicomplexa are susceptible to certain antibiotics that usually only affect bacteria • Apicomplexa have a subcellular structure in the vicinity of the nucleus that is surrounded ...
... • Apicomplexa appear to have three genomes • Apicomplexa are susceptible to certain antibiotics that usually only affect bacteria • Apicomplexa have a subcellular structure in the vicinity of the nucleus that is surrounded ...
1-2 - FaPGenT
... • The DNA in living cells is contained within large structures termed chromosomes • Human cells have a total of 46 chromosomes • Each chromosome is a complex of DNA and proteins • An average human chromosome contains – More than a 100 million nucleotides – about 1,000 different genes ...
... • The DNA in living cells is contained within large structures termed chromosomes • Human cells have a total of 46 chromosomes • Each chromosome is a complex of DNA and proteins • An average human chromosome contains – More than a 100 million nucleotides – about 1,000 different genes ...
A Novel Chimeric Low-Molecular-Weight Glutenin
... (DDMMUU) were isolated and characterized. Sequence structures showed that the 4 LMW-m-type genes, assigned to the M genome of Ae. neglecta, displayed a high homology with those from hexaploid common wheat. The novel chimeric gene, designed as AjkLMW-i, was isolated from both Ae. kotschyi and Ae. juv ...
... (DDMMUU) were isolated and characterized. Sequence structures showed that the 4 LMW-m-type genes, assigned to the M genome of Ae. neglecta, displayed a high homology with those from hexaploid common wheat. The novel chimeric gene, designed as AjkLMW-i, was isolated from both Ae. kotschyi and Ae. juv ...
Teacher Materials
... occur in either order (A-T, T-A, C-G, G-C). The bases are thus in only four different combinations in relation to their connections with the ladder uprights, although they form many different sequences along the DNA uprights. Each base represents a “code letter .” A “code word” or codon is formed by ...
... occur in either order (A-T, T-A, C-G, G-C). The bases are thus in only four different combinations in relation to their connections with the ladder uprights, although they form many different sequences along the DNA uprights. Each base represents a “code letter .” A “code word” or codon is formed by ...
Deletion loops in polytene chromosomes
... Most SINES and LINES in human genome are defective TEs Nonautonomous elements – need activity of nondeleted copies of same TE for movement Autonomous elements – move by themselves ...
... Most SINES and LINES in human genome are defective TEs Nonautonomous elements – need activity of nondeleted copies of same TE for movement Autonomous elements – move by themselves ...
Presentation
... Understanding users, system customization and training is as important as new technology ...
... Understanding users, system customization and training is as important as new technology ...
FREE Sample Here
... 41) What is another term for a biological catalyst? Answer: enzyme Section: 1.3 42) Research dealing with which human blood disorder was instrumental in linking the genotype to a specific phenotype, and what conclusion was reached? Answer: The work on sickle-cell anemia was instrumental in showing t ...
... 41) What is another term for a biological catalyst? Answer: enzyme Section: 1.3 42) Research dealing with which human blood disorder was instrumental in linking the genotype to a specific phenotype, and what conclusion was reached? Answer: The work on sickle-cell anemia was instrumental in showing t ...
Document
... series of reversals to transform one into another • Input: Permutations p and s • Output: A series of reversals r1,…rt transforming p into s, such that t is minimum • t - reversal distance between p and s • d(p, s) = smallest possible value of t, given p, s ...
... series of reversals to transform one into another • Input: Permutations p and s • Output: A series of reversals r1,…rt transforming p into s, such that t is minimum • t - reversal distance between p and s • d(p, s) = smallest possible value of t, given p, s ...
Bioinformatics Seminar 13/11/07
... gabos -afile refFlat.txt -genome mm9 -seqrange 4,482,560-4,483,185 -chr 1 -pre 420 -post 420 –fastaonly >my_results.fa Options can be in any order. Output can be redirected to a file as shown. A file of gene names could be used as input instead of a chromosome sequence range. gabos –help lists all o ...
... gabos -afile refFlat.txt -genome mm9 -seqrange 4,482,560-4,483,185 -chr 1 -pre 420 -post 420 –fastaonly >my_results.fa Options can be in any order. Output can be redirected to a file as shown. A file of gene names could be used as input instead of a chromosome sequence range. gabos –help lists all o ...