* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download workshop-1
Cre-Lox recombination wikipedia , lookup
Gene regulatory network wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Gene desert wikipedia , lookup
Exome sequencing wikipedia , lookup
Gene expression profiling wikipedia , lookup
Gene expression wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Whole genome sequencing wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Molecular ecology wikipedia , lookup
Community fingerprinting wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Point mutation wikipedia , lookup
Genomic library wikipedia , lookup
Non-coding DNA wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Molecular evolution wikipedia , lookup
Genome evolution wikipedia , lookup
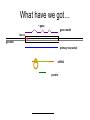
Bioinformatics Workshops 1 & 2 1. use of public database/search sites - range of data and access methods - interpretation of search results - understanding the meaning & effect of search (e.g. BLAST) parameters 2. functional analysis of single sequences - i.e. how to work out what your unknown protein might be doing - complex searches for (e.g.) patterns of motifs & secondary structure elements Workshop 1. overall survey of data Main data axes Biological origin of sequences Main Portals Genes vs.loci Mutation between species -> orthologs Mutation between duplications -> domains Database searches vs. genome browsers Finding similar sequences BLAST, et al E-values! Search methods – 2D vs. 3D Random sequences Search methods – similarity vs. models vs. comparative Using Public Data Resources • There is (are!) data out there • There are methods out there • Quite often they are combined – BLAST searches of sequence databases Notes… • Sequence databases – Entrez queries… • • • • Genome browsers/databases Regulatory Elements SNPs Functional Sequence Models (PFam domains, etc.) • Expression Data – Array data – in situ data Notes II • Blast parameters – Low complexity: frameshifted cDNA – miRNAs vs genome – morpholinos for other genes – -q-2 for EST vs EST alignments – Entrez queries What have we got… ~ gene gene model locus genome primary transcript mRNA protein Derivative Sequences mRNA clone into cDNA library 5’ EST Single pass sequence from each end of the clone 3’ EST cDNA sequence Multiple pass sequencing over whole length of the clone Initial Growth of Databases • Lots of ESTs were generated • Some clones were selected for full-insert sequencing -> cDNAs • cDNAs were translated to yield presumed protein sequences Then Came Genomes • With increasing larger fragments of genomic sequence came the ability to align cDNAs to create gene models • And then to apply our understanding of exon/intron structure to predict theoretical genes… Introns and Exons mRNA CTACCATCCATGCTAACCATTCTACCATTTTATACTCATGCAACGGACCGTAGCGTAGTCGCTTAGCATCCTTTATAACTGGCTA gene model genome exon intron exon intron exon splice sites CTACCATCCATGCTAACCATTCTAC CATTTTATACTCATGCAACGGACCGT AGCGTAGTCGCTTAGCATCCTTTATAACTGGCTA CTACCATCCATGCTAACCATTCTACGTAAGTCATCTATATCAATATTATTTCAGCATTTTATACTCATGCAACGGACCGTGTCAGTATTACAGAGCGTAGTCGCTTAGCATCCTTTATAACTGGCTA GTAAG. .TTTCAG donor acceptor Gene Predictions Given: - coding sequence must run from ATG – STOP codon in-frame - introns GT. . . . . . AG can be spliced out Also take a statistical approach: - coding and non-coding sequence are slightly different in composition - some ‘possible’ splice sites are more likely than others scan genomic sequence … . . .CGTCGTATGGCTTCGATGTAGTACATCGGATCGGTATGGAATCATTTCAGTCGCTAGCTAGCCTAACGTATATAGCTAGGTAAGACTA. . . . .CGTCGTATGGCTTCGATTCGCTAGCTAGCCTAACGTATATAGCTAGGTAAGACTA. .CGTCGTATGGCTTCGATGTAGTACATCGGATCGGTATGGAATCATTTCAGTCGCTAGCTAGCCTAACGTATATAGCTAGGTAAGACTA. . . . .CGTCGTATGGCTTCGATGTAGTACATCGGATCGGTATGGAATCATTTCAGTCGCTAGCTAGCCTAACGTATATAGCTAGGTAAGACTA. . . . .CGTCGTATGGCTTCGATGTAGTACATCGGATCGTCGCTAGCTAGCCTAACGTATATAGCTAGGTAAGACTA. . . most likely gene model . . .CGTCGTATGGCTTCGATGTAGTACATCGGATCGGTATGGAATCATTTCAGTCGCTAGCTAGCCTAACGTATATAGCTAGGTAAGACTA. . Supporting Evidence! exons: 1 2 3 4 gene model genome EST evidence We note that even though there is good evidence for the existence of all four exons, there is no evidence that all the exons would appear on a real transcript. An alternative transcript, skipping exon 3, would be plausible, if a little unlikely. This gets less ambiguous as more ESTs are available, and clones are sequenced at both ends (which helps put distant exons into the same transcripts), and eventually full-length transcript sequences are available. So What’s in the Databases Now? • At NCBI – 15,000,000 EST sequences – 3,329,110 non-redundant DNA sequences (excluding ESTs, etc.) – 2,693,904 non-redundant translated coding sequences – 954,378 Protein Reference Sequences sequences (RefSeq) • But the majority of RefSeq may be translations of theoretical transcripts… Main Data Axes • Europe: EBI/EMBL – Swiss-Prot/Trembl/Ensembl/UniProt • US: NIH/NCBI – GenBank/UniGene/RefSeq/Entrez • Japan: DNA Data Bank of Japan – National Institute of Genetics Synchronisation… You submit a sequence ATCGATCGATCATAGTATGCTAGCTGCTA GenBank EMBL BC009638.1 ATCGATCGATCATAGTATGCTAGCTGCTA DDBJ Sequences, Accession Numbers and Genes NM_001015922.1 gi=62860271 GATCGTTCGATTAGCTAGGGACACCACCGATCGATATGACCACAAAAA NM_001015922.2 gi=62860589 GACCGTTCGATTAGCTAGGGACACCACCGATCGATATGACCACAAAAA BC009638.1 gi=16307106 GTTCGATTAGCTAGGGACACCACCGATCGATATGACCACAAAA Main Data Portals • • • • • NCBI Entrez Databases ExPASy Proteomics Server DNA Data Bank of Japan DDBJ EBI Ensembl Genome Browser Santa Cruz Genome Browser