* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download MolecularModelingDru..
Artificial gene synthesis wikipedia , lookup
Magnesium transporter wikipedia , lookup
Ligand binding assay wikipedia , lookup
Expression vector wikipedia , lookup
Gene expression wikipedia , lookup
Paracrine signalling wikipedia , lookup
Point mutation wikipedia , lookup
Multi-state modeling of biomolecules wikipedia , lookup
Biochemistry wikipedia , lookup
Interactome wikipedia , lookup
Metalloprotein wikipedia , lookup
Signal transduction wikipedia , lookup
Protein purification wikipedia , lookup
Western blot wikipedia , lookup
Drug discovery wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Proteolysis wikipedia , lookup
Clinical neurochemistry wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Molecular Modeling and
Drug Discovery
Judith Klein-Seetharaman
Assistant Professor
Department of Pharmacology
University of Pittsburgh School of Medicine
Background
• View of living organisms
as molecular circuitry:
– Molecular circuitry =
biochemical processes,
that form and recycle
molecules in a
coordinated and
balanced fashion
– intended modes of
operation = healthy
state
– aberrant modes of
operation = disease
state
• Diagnosis:
– identify the molecular
basis of disease
• Therapy:
– guide biochemical
circuitry back to healthy
state
Information Sources
• New technology generates
massive amounts of data
(often stored in publicly
accessible databases):
Genomics and Proteomics
– Protein and DNA
sequences / Whole
genome sequences
– Protein structure data
– Protein pathways and
networks
– Protein interaction data
– Expression data
Genomics - Proteomics
Mapping Sequence to Protein Structure and Dynamics
Primary Sequence
MNGTEGPNFY
PLNYILLNLA
KPMSNFRFGE
HFIIPLIVIF
SDFGPIFMTI
VPFSNKTGVV
VADLFMVFGG
NHAIMGVAFT
FCYGQLVFTV
PAFFAKTSAV
Folding
3D Structure
RSPFEAPQYY
FTTTLYTSLH
WVMALACAAP
KEAAAQQQES
YNPVIYIMMN
LAEPWQFSML
GYFVFGPTGC
PLVGWSRYIP
ATTQKAEKEV
KQFRNCMVTT
AAYMFLLIML
NLEGFFATLG
EGMQCSCGID
TRMVIIMVIA
LCCGKNPLGD
GFPINFLTLY
GEIALWSLVV
YYTPHEETNN
FLICWLPYAG
DEASTTVSKT
VTVQHKKLRT
LAIERYVVVC
ESFVIYMFVV
VAFYIFTHQG
ETSQVAPA
Genomics - Proteomics
Mapping Sequence to Protein Structure, Dynamics and Function
Primary Sequence
MNGTEGPNFY
PLNYILLNLA
KPMSNFRFGE
HFIIPLIVIF
SDFGPIFMTI
VPFSNKTGVV
VADLFMVFGG
NHAIMGVAFT
FCYGQLVFTV
PAFFAKTSAV
Folding
3D Structure
RSPFEAPQYY
FTTTLYTSLH
WVMALACAAP
KEAAAQQQES
YNPVIYIMMN
LAEPWQFSML
GYFVFGPTGC
PLVGWSRYIP
ATTQKAEKEV
KQFRNCMVTT
AAYMFLLIML
NLEGFFATLG
EGMQCSCGID
TRMVIIMVIA
LCCGKNPLGD
GFPINFLTLY
GEIALWSLVV
YYTPHEETNN
FLICWLPYAG
DEASTTVSKT
VTVQHKKLRT
LAIERYVVVC
ESFVIYMFVV
VAFYIFTHQG
ETSQVAPA
Challenge 1:
Disease Causing Mutations
Primary Sequence
MNGTEGPNFY
PLNYILLNLA
KPMSNFRFGE
HFIIPLIVIF
SDFGPIFMTI
VPFSNKTGVV
VADLFMVFGG
NHAIMGVAFT
FCYGQLVFTV
PAFFAKTSAV
RSPFEAPQYY
FTTTLYTSLH
WVMALACAAP
KEAAAQQQES
YNPVIYIMMN
LAEPWQFSML
GYFVFGPTGC
PLVGWSRYIP
ATTQKAEKEV
KQFRNCMVTT
AAYMFLLIML
NLEGFFATLG
EGMQCSCGID
TRMVIIMVIA
LCCGKNPLGD
GFPINFLTLY
GEIALWSLVV
YYTPHEETNN
FLICWLPYAG
DEASTTVSKT
Folding
3D Structure
Complex function within
network of proteins
Disease
VTVQHKKLRT
LAIERYVVVC
ESFVIYMFVV
VAFYIFTHQG
ETSQVAPA
Challenge 1:
Non-disease causing mutations
Primary Sequence
MNGTEGPNFY
PLNYILLNLA
KPMSNFRFGE
HFIIPLIVIF
SDFGPIFMTI
VPFSNKTGVV
VADLFMVFGG
NHAIMGVAFT
FCYGQLVFTV
PAFFAKTSAV
RSPFEAPQYY
FTTTLYTSLH
WVMALACAAP
KEAAAQQQES
YNPVIYIMMN
LAEPWQFSML
GYFVFGPTGC
PLVGWSRYIP
ATTQKAEKEV
KQFRNCMVTT
AAYMFLLIML
NLEGFFATLG
EGMQCSCGID
TRMVIIMVIA
LCCGKNPLGD
GFPINFLTLY
GEIALWSLVV
YYTPHEETNN
FLICWLPYAG
DEASTTVSKT
Folding
3D Structure
Complex function within
network of proteins
Normal
VTVQHKKLRT
LAIERYVVVC
ESFVIYMFVV
VAFYIFTHQG
ETSQVAPA
Challenge 1
How can we distinguish functional from nonfunctional protein sequences?
Needed: sequence to structure and function mapping
Challenge 2:
Which protein is a drug target?
Challenge 3:
How to design a drug in the absence of a structure?
Drug Target:
?
Challenge 4:
Drug action, efficacy and side effects?
Drug Target:
Challenges for Bioinformatics in
Drug Discovery
1. How can we distinguish functional from non-functional
protein sequences?
2. Which protein is a drug target?
3. How to design a drug in the absence of a structure?
4. Understanding drug action, efficacy and side effects
Fundamental Scientific Challenge:
Mapping the relationship between genome sequence and
protein structures, dynamics and functions in complex
cellular environments
Meaning for drug discovery
• If one could predict the structure of proteins from
sequence, one could discover new drugs at a
fast pace
• If one could predict the relationship between
isozyme and tissue expression, one could
design drugs specific to certain tissues
• If one could predict the interactions of proteins in
different protein networks, one could interpret
complex data such as animal models
• If one could…
Mapping relationships: 7 hierarchical layers
Layer 5. Predicting functional
• Layer 1. Sequencing support
structures (DNA - RNA – (physical mapping, fragment assembly
outcome: raw genome sequence)
proteins - lipids • Layer 2. DNA sequence analysis
carbohydrates)
1. Gene finding
1. Homology modeling
2. non-coding sequences
2. ab initio
3. regulatory sequences finding
3. templates
4. orthologous and paralogous sequences
4. partial information
5. Evolution
1. overall architecture
• Layer 3. Protein sequence analysis
2. binding pocket
1. homology detection
3. protein backbone
2. alignment
Layer 6. Molecular
3. functional annotation
interactions
4. cellular localization
(Protein-ligand, -protein, -DNA, • Layer 4. From linear sequence to threeRNA, -lipid, -carbohydrate)
dimensional shapes
Layer 7. Gene expression,
– conformational space
metabolic and regulatory
– models for protein (mis)folding
networks
– discriminating structures
– conformational ambiguity
Specific Challenges for
Bioinformatics in Drug Discovery
• Data needs to be organized, mined and
visualized to allow scientific discovery
• Linking variety of databases
• Linking the different layers
• Interpretation of data
• Drug discovery
Outline Drug Discovery Approach
• use the information in the databases and infer
information that is not provided directly by genomics and
proteomics data: higher level information
=> piece together all available information
- to get detailed picture of a molecular process (or
disease)
- to identify new protein targets
- to develop drugs
• based on chemical similarity of known drugs
• rational (structure-based) drug design interactively
on computer screen
• molecular docking (automatic, systematic
computer-based prediction of structure and
binding affinity of complex)
• high-throughput screening and combinatorial
chemistry
Molecular modeling in drug discovery
I.
Two case studies for sequence to structure
mapping:
–
–
II.
Small changes in protein sequence cause dramatic
difference in drug binding: COX inhibitors
Large changes in protein sequence still maintain
similar structure: G protein coupled receptors
Protein Structure Prediction
III. Ligand Docking to Protein Structures
Molecular modeling in drug discovery
I.
Two case studies for sequence to structure
mapping:
–
–
II.
Small changes in protein sequence cause dramatic
difference in drug binding: COX inhibitors
Large changes in protein sequence still maintain
similar structure: G protein coupled receptors
Protein Structure Prediction
III. Ligand Docking to Protein Structures
Case study COX
A Wonder Drug: What is the most commonly-taken drug today?
It is an effective painkiller.
It reduces fever and inflammation when the body gets overzealous in
its defenses against infection and damage.
It slows blood clotting, reducing the chance of stroke and heart attack
in susceptible individuals.
It may be an effective addition to the fight against cancer.
Aspirin has been used professionally for a century, and
traditionally since ancient times. A similar compound
found in willow bark, salicylic acid, has a long history of
use in herbal treatment. But only in the last few decades
have we understood how aspirin works, and how it might
be improved
http://www.rcsb.org/pdb/molecules/pdb17_1.html
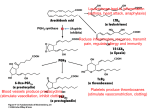
Prostaglandins
As you might expect from a drug with such diverse actions, aspirin blocks a
central process in the body: Aspirin blocks the production of
prostaglandins, key hormones that are used to carry local messages.
Unlike most hormones, which are produced in specialized glands and then
delivered throughout the body by the blood, prostaglandins are created by cells
and then act only in the surrounding area before they are broken down.
Prostaglandins control many of these neighborhood processes, including the
constriction of muscle cells around blood vessels, aggregation of platelets
during blood clotting, and constriction of the uterus during labor.
Prostaglandins also deliver and strengthen pain signals and induce
inflammation.
When aspirin blocks production of prostaglandins, the normal messages are
not delivered, so we don't feel the pain and don't launch an inflammation
response.
These many different processes are all controlled by different prostaglandins,
but all created from a common precursor molecule.
http://www.rcsb.org/pdb/molecules/pdb17_1.html
Arachidonic Acid and COX
What does COX do?
COX = Cyclooxygenase (PDB entry 1prh) performs the first step in the
creation of prostaglandins from a common fatty acid: It adds two
oxygen molecules to arachidonic acid, beginning a set of reactions.
http://www.rcsb.org/pdb/molecules/pdb17_1.html
Two different active sites,
collectively prostaglandin
synthase: 1, the
cyclooxygenase active site
discussed; 2, is has an entirely
separate peroxidase site,
which is needed to activate the
heme groups that participate in
the cyclooxygenase reaction.
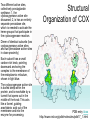
Structural
Organization of COX
Dimer of identical subunits (two
cyclooxygenase active sites
and two peroxidase active sites
in close proximity)
Each subunit has a small
carbon-rich knob, pointing
downward anchoring the
complex to the membrane of
the endoplasmic reticulum,
shown in light blue.
The cyclooxygenase active site
is buried deep within the
protein, and is reachable by a
tunnel that opens out in the
middle of the knob. This acts
like a funnel, guiding
arachidonic acid out of the
membrane and into the
enzyme for processing.
PDB entry 4cox
http://www.rcsb.org/pdb/molecules/pdb17_1.html
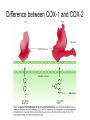
Why is there a COX-1 and COX-2?
COX-1 and COX-2 are made for different purposes.
COX-1 is built in many different cells to create
prostaglandins used for basic housekeeping messages
throughout the body.
COX-2 is built only in special cells and is used for signaling
pain and inflammation.
Aspirin attacks both. Since COX-1 is targeted, aspirin can
lead to unpleasant complications, such as stomach
bleeding.
Needed: specific compounds that block just COX-2, leaving
COX-1 to perform its essential jobs. These drugs are
selective pain-killers and fever reducers, without the
unpleasant side-effects.
How would you design a drug that
only blocks COX-2 and not COX-1?
Structure-based drug design
• Compare the structures of COX-1 and
COX-2
• Identify differences that could be exploited
• Need to know the mechanism of COX
inhibition
Mechanism of COX inhibition?
Ser530 is not a catalytic
residue.
But it is located in the
tunnel that allows entry of
arachidonic acid to the
active site.
Aspirin sterically blocks the
binding of arachidonic acid in
the cyclooxygenase active
site.
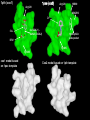
Structural Differences COX-1 & COX-2
1pxx (cox2)
1pth (cox1)
Arg120
Arg120
HIS90
ARG513
DIF
SAL
Ile (COX-1) Val523 (COX-2)
Hydrophilic
side pocket
BRM
Tyr385
Tyr385
Hydrophilic Side Pocket
HIS90
ARG513
Hydrophilic
side pocket
Difference between COX-1 and COX-2
Structural Differences Cox1/2
Rasmol
His90
Arg513
Summary COX Case Study
• Being able to model the effect of small
changes in sequence (isoforms) is
essential for drug development
Molecular modeling in drug discovery
I.
Two case studies for sequence to structure
mapping:
–
–
II.
Small changes in protein sequence cause dramatic
difference in drug binding: COX inhibitors
Large changes in protein sequence still maintain
similar structure: G protein coupled receptors
Protein Structure Prediction
III. Ligand Docking to Protein Structures
Structure Analysis
• How does one determine structures?
– Experimentally (X-ray, NMR)
– Computationally (ab initio, Rosetta, threading,
homology models)
• How does one access structures?
– Pdb
– SCOP/CATH
• How does one analyze structures?
– Visualization programs (chime, rasmol, molmol,
Insight etc.)
Modeling Methods and Relation to
Sequence Similarity
A. When no information but sequence and physical principles are used
= ab initio structure prediction (Blue Gene IBM )
B. When other information is used ("ab initio" methods that use pdb
information)
Common features: "fold recognition“, requires a method for evaluating
the compatibility of a given sequence with a given folding pattern
1. 3D profiles
2. Rosetta: conformations from short segments in pdb
3. Including experimental structural constraints
4. Threading (=sequence-structure alignment),
5. Inverse threading and folding experiments
a. using short-range information
b. using short- and long-range information
6. Predicting structural class only
7. Predicting active site only
8. Predicting protein-protein interaction sites
9. Predicting surface shape?
Modeling Methods Continued
C. When a template with known structure must be available: homology
modeling
D. Modeling structures based on experimental data
Both NMR and X-ray underdetermine the protein structure. To solve a
structure one must minimize a combination of the deviation from the
experimental data and the conformational energy:
a. NMR (set of constraints on distances and angles)
b. X-ray crystallography (Fourier transform of the electron density)
Evaluating structure prediction
• Use rmsd to known structures - defines
structural similarity
• Critical Assessment of Structure
Predictions (CASP) competitions
• EVA, EVA submits sequences
automatically to different prediction
servers shortly before structures are
published in pdb
Homology Modeling
• Database searching for homologous proteins
( Blast the query sequence towards the pdb database )
• Alignment (Pairwise/ Multiple Alignments)
– needs minimum 30% sequence identity, but to be useful
usually need 40-50%
– note that ~30% of genomes have sequence identity of 20%
• Model Building
– Modeller , Composer etc
• Model Refinement and Evaluation
– Joy
– Procheck etc
BLAST is a heuristic search
method that seeks words of
length W (default = 3 in
blastp) that score at least T
when aligned with the query
and scored with a
substitution matrix. Words in
the database that score T or
greater are extended in both
directions in an attempt to
fina a locally optimal
ungapped alignment or HSP
(high scoring pair) with a
score of at least S or an E
value lower than the
specified threshold. HSPs
that meet these criteria will
be reported by BLAST,
provided they do not exceed
the cutoff value specified for
number of descriptions
and/or alignments to report.
BLAST (Basic Local
Alignment Search Tools)
BLOSUM62 Substitution Scoring Matrix. The BLOSUM
62 matrix shown here is a 20 x 20 matrix of which a section
is shown here in which every possible identity and
substitution is assigned a score based on the observed
frequencies of such occurences in alignments of related
proteins. Identities are assigned the most positive scores.
Frequently observed substitutions also receive positive
scores and seldom observed substitutions are given
negative scores.
Scoring matrices
The PAM family PAM matrices are based on global
alignments of closely related proteins. The PAM1 is the
matrix calculated from comparisons of sequences with no
more than 1% divergence. Other PAM matrices are
extrapolated from PAM1.
The BLOSUM family BLOSUM matrices are based on
local alignments. BLOSUM 62 is a matrix calculated from
comparisons of sequences with no less than 62%
divergence. All BLOSUM matrices are based on observed
alignments; they are not extrapolated from comparisons of
closely related proteins. BLOSUM 62 is the default matrix in
BLAST 2.0. Though it is tailored for comparisons of
moderately distant proteins, it performs well in detecting
closer relationships. A search for distant relatives may be
more sensitive with a different matrix.
The relationship between BLOSUM and PAM
substitution matrices. BLOSUM matrices with higher
numbers and PAM matrices with low numbers are both
designed for comparisons of closely related sequences.
BLOSUM matrices with low numbers and PAM matrices
with high numbers are designed for comparisons of
distantly related proteins. If distant relatives of the query
sequence are specifically being sought, the matrix can be
tailored to that type of search.
http://www.ncbi.nlm.nih.gov/Education/
Blast Result COX-1 vs. COX-2
Score =
Query: 13
745 bits (1924), Expect = 0.0Identities = 343/560 (61%),
Positives = 433/560 (77%), Gaps = 1/560 (0%)
GLSQAANPCCSNPCQNRGECMSTGFDQYKCDCTRTGFYGENCTTPEFLTRIKLLLKPTPN 72
G
NPCC
PCQ++G C+
G D+Y+CDCTRTG+ G NCT PE
T ++
L+P+P+
Sbjct: 4
GAPAPVNPCCYYPCQHQGICVRFGLDRYQCDCTRTGYSGPNCTIPEIWTWLRTTLRPSPS 63
Query: 73
TVHYILTHFKGVWNIVNNIPFLRSLIMKYVLTSRSYLIDSPPTYNVHYGYKSWEAFSNLS 132
Sbjct: 64
FIHFLLTHGRWLWDFVN-ATFIRDTLMRLVLTVRSNLIPSPPTYNIAHDYISWESFSNVS 122
+H++LTH + +W+ VN
F+R
+M+ VLT RS LI SPPTYN+ + Y SWE+FSN+S
Query: 133 YYTRALPPVADDCPTPMGVKGNKELPDSKEVLEKVLLRREFIPDPQGSNXXXXXXXXXXX 192 Query: 373 WHPLLPDTFNIEDQEYSFKQFLYNNSILLEHGLTQFVESFTRQIAGRVAGGRNVPIAVQA 432
YYTR LP V
DCPTPMG KG K+LPD++ +
+ LLRR+FIPDPQG+N
WHPL+PD+F +
Q+YS++QFL+N S+L+++G+
V++F+RQ AGR+ GGRN+
+
Sbjct: 123 YYTRILPSVPRDCPTPMGTKGKKQLPDAEFLSRRFLLRRKFIPDPQGTNLMFAFFAQHFT 182 Sbjct: 363 WHPLMPDSFRVGPQDYSYEQFLFNTSMLVDYGVEALVDAFSRQPAGRIGGGRNIDHHILH 422
Query: 193 XXXXXXDHKRGPGFTRGLGHGVDLNHIYGETLDRQHKLRLFKDGKLKYQVIGGEVYPPTV 252 Query: 433 VAKASIDQSREMKYQSLNEYRKRFSLKPYTSFEELTGEKEMAAELKALYSDIDVMELYPA 492
K GPGFT+ LGHGVDL HIYG+ L+RQ++LRLFKDGKLKYQ++ GEVYPP+V
VA
I +SR ++ Q
NEYRKRF +KPYTSF+ELTGEKEMAAEL+ LY DID +E YP
Sbjct: 183 HQFFKTSGKMGPGFTKALGHGVDLGHIYGDNLERQYQLRLFKDGKLKYQMLNGEVYPPSV 242 Sbjct: 423 VAVDVIKESRVLRLQPFNEYRKRFGMKPYTSFQELTGEKEMAAELEELYGDIDALEFYPG 482
Query: 253 KDTQVEMIYPPHIPENLQFAVGQEVFGLVPGLMMYATIWLREHNRVCDILKQEHPEWGDE 312 Query: 493 LLVEKPRPDAIFGETMVELGAPFSLKGLMGNPICSPQYWKPSTFGGEVGFKIINTASIQS 552
++
V M YP
IP
Q AVGQEVFGL+PGLM+YATIWLREHNRVCD+LK EHP WGDE
LL+EK
P++IFGE+M+E+GAPFSLKGL+GNPICSP+YWK STFGGEVGF ++ TA+++
Sbjct: 243 EEAPVLMHYPRGIPPQSQMAVGQEVFGLLPGLMLYATIWLREHNRVCDLLKAEHPTWGDE 302 Sbjct: 483 LLLEKCHPNSIFGESMIEMGAPFSLKGLLGNPICSPEYWKASTFGGEVGFNLVKTATLKK 542
Query: 313 QLFQTSRLILIGETIKIVIEDYVQHLSGYHFKLKFDPELLFNQQFQYQNRIASEFNTLYH 372 Query: 553 LICNNVKGCPFTSFNVQDPQ 572
QLFQT+RLILIGETIKIVIE+YVQ LSGY
+LKFDPELLF
QFQY+NRIA EFN LYH
L+C N K CP+ SF+V DP+
Sbjct: 303 QLFQTARLILIGETIKIVIEEYVQQLSGYFLQLKFDPELLFGAQFQYRNRIAMEFNQLYH 362 Sbjct: 543 LVCLNTKTCPYVSFHVPDPR 562
Model Building
• Modeller (freeware,
http://www.salilab.org/modeller/modeller.html)
• Spdbviewer Swissmodel–module (freeware,
http://us.expasy.org/spdbv/)
• Composer (module of InsightII, commercial
version of Modeller)
Model Building Principles
• Sequentially go from amino acid position to next
position
– if same amino acid, copy the coordinates
– If different amino acid, if the new amino acid has
atoms in common with the template, those atoms will
be copied, and the rest are computed
• At every step, check for steric clashes with
previous amino acids
– Minimization allowing the position of new amino acid
to change
– Only at the final stage, bond energy is minimized
Model Refinement and Evaluation
http://cgat.ukm.my/spores/Predictory/evaluation.html
• Verify3D (based on
surface accessibility)
• Procheck (based on
phi/psi angle, rmsd
deviations)
• Joy (based on
secondary structure
assignments)
• WHAT IF (bond length,
bond angles, chi values,
etc.)
WHAT IF Checklist
A WHAT IF check report: what does it mean?
General points
Administrative checks
Nomenclature
Chain name
Weights (occupancy)
Missing atoms and C-terminal oxygens
Symmetry
Consistency
Cell conventions
Matthews' Coefficient
Higher symmetry
Non crystallographic symmetry
Geometry
Chirality
Bond lengths
Bond angles
Torsion Angles: "Evaluation"; "Ramachandran"; "omega"; "Chi1/2"
Rings and planarity: "Planarity"; "Proline Puckering"
Structure
Inside/outside profile
Bumps
Packing quality
Backbone: "number of hits"; "backbone normality"; "peptide flips"
Sidechain rotamers
Water molecules: "floating clusters"; "symmetry relations"
B-factors: "average"; "low B-factors"; "B-factor distribution"
Hydrogen bonds: "Flip check"; "HIS assignments"; "Unsatisfied"
Collection of homology models
• MODBASE
– uses PSI-BLAST plus MODELLER to model
and stores coordinates in this database
• SWISS-MODEL
– automatic structure prediction
Play with homology models
• www.cs.cmu.edu/~blmt/Seminar/SeminarMaterials/COX
COX 2 Modelling :
Template structure : 1PTH.pdb (cox1 in ovis aries)
query seq:sequence of 1PXX.pdb (cox2 in mus musculus)
model generated using modeller: 2cox.pdb
COX 1 Modelling:
Template structure : 1PXX.pdb (cox2 in mus musculus)
query seq:sequence of 1PTH.pdb (cox1 in ovis aries)
model generated using modeller: 1cox.pdb
• Rasmol is also in this directory, just click on the raswin
icon to start program
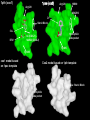
1pxx (cox2)
1pth (cox1)
Arg120
Arg120
HIS90
ARG513
DIF
SAL
Ile (COX-1) Val523 (COX-2)
Hydrophilic
side pocket
BRM
Tyr385
Tyr385
cox1 model based
on 1pxx template
Cox2 model based on 1pth template
1pxx (cox2)
1pth (cox1)
Arg120
Arg120
HIS90
ARG513
DIF
Steric Block
SAL
BRM
Hydrophilic
side pocket
Ile (COX-1) Val523 (COX-2)
Tyr385
Tyr385
cox1 model based
on 1pxx template
Cox2 model based on 1pth template
Steric Block
Hydrophilic
side pocket
Molecular modeling in drug discovery
I.
Two case studies for sequence to structure
mapping:
–
–
II.
Small changes in protein sequence cause dramatic
difference in drug binding: COX inhibitors
Large changes in protein sequence still maintain
similar structure: G protein coupled receptors
Protein Structure Prediction
III. Ligand Docking to Protein Structures
Protein-ligand docking
• First (if structure is known) or second (after structure
prediction) step in a drug design project: find a lead
structure (=small molecule which binds to a given target)
• docking problem - predicting the energetically most
favorable complex between a protein and a putative drug
molecule
• For a given protein structure, one can apply docking
algorithms to virtually search through the space
• 2 questions:
1. what does the protein-ligand complex look like
2. what is the affinity with respect to other candidates?
What makes the docking problem
hard to solve?
1. Scoring problem
– = calculating binding affinity given a protein-ligand
complex
– no general scoring function is available
2. Large number of degrees of freedom
– most important degrees of freedom:
1.
2.
3.
4.
5.
relative orientation of the two molecules
conformation of the ligand
protein conformation
water molecules can be between ligand and protein
protonation state
Types of Docking Problems
1. Macromolecular docking
• = two macromolecules are docked, such as protein and
DNA, or protein and protein
• large contact area
• molecules have fixed overall shape
• => methods based on geometric properties like shape
complementarities alone can be efficiently used to create
energetically favorable complexes
2. Small molecule docking
• = a small molecule is docked to a macromolecule
• ligand is typically not fixed in shape (as opposed to
macromolecular docking)
• typical ligand size has 5-12 rotatable bonds
• often fragments of ligand are used for modeling, eg.
combinatorial libraries are docked by combining
placement for individual building blocks of the library
Steps in Molecular Docking
1. Find a set of compounds to start with
- e.g. from inspecting known ligands for a protein (e.g.
substrate in an enzyme)
2. compounds from a screening experiment of a
combinatorial library (in which there is usually a
molecular fragment that is common between all
molecules of the library, the core, and the fragments
attached to the core are R-groups)
3. compounds from a filtering experiment using other
software
4. from varying other lead structures or known ligands
5. virtual screening using a fast docking algorithm (typically
from a million molecules)
6. de novo design using fragments of compounds
=> get several hundred to thousands of ligands to start
with
Docking Methods
• Rigid-body docking algorithms
– Historically the first approaches.
– Protein and ligand are held fixed in conformational
space which reduces the problem to the search for
the relative orientation fo the two molecules with
lowest energy.
– All rigid-body docking methods have in common that
superposition of point sets is a fundamental subproblem that has to be solved efficiently:
– Superposition of point sets: minimize the RMSD
• Flexible ligand docking algorithms
– most ligands have large conformational spaces with
several low energy states
http://www-2.cs.cmu.edu/~blmt/Seminar/SeminarMaterials/interactions.html
Clique-search based approaches
• = matching characteristic features of the two molecules
• use a graph to search for compatible matches: the
vertices of the graph are all possible matches and edges
connect pairs of vertices representing compatible
matches
• compatibility = distance compatibility with in a fixed
tolerance epsilon
• The matches (p1,l1), (p2,l2) are distance-compatible if
|d(p1,p2)-d(l1,l2)| < epsilon
• Example: DOCK program
DOCK
= today most widely used molecular docking program
• starting with the molecular surface of the protein , a set of spheres is created
inside the active sties
• the spheres represent the volume which could be occupied by the ligand:
VOLUME is the feature used for matching
• ligand is represented by spheres inside the ligand
For more information: http://www.cmpharm.ucsf.edu/kuntz/dock_demo.html
Molecular modeling in drug discovery
I.
Two case studies for sequence to structure
mapping:
–
–
II.
Small changes in protein sequence cause dramatic
difference in drug binding: COX inhibitors
Large changes in protein sequence still maintain
similar structure: G protein coupled receptors
Protein Structure Prediction
III. Ligand Docking to Protein Structures
G Protein Coupled Receptors
C
Cytoplasmic Domain
Transmembrane
Domain
1
2
3
4
5
6
7
Extracellular Domain
N
• Largest family of cell surface receptors
• >8000 sequences known
• 60% of all known drugs target GPCR
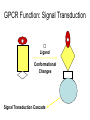
GPCR Function: Signal Transduction
Ligand
Conformational
Changes
Signal Transduction Cascade
GPCR Family and Their Ligands
Class A: Rhodopsin-like Family
Opsins, Odorants, Monoamines, Lipid messengers, Purines, Neuropeptides, Peptide
hormones (e.g. platelet activating factor, gonadotropin -releasing hormone, th yrotropin
releasing hormone & melatonin), Glycoprotein hormones, Chemokines, Proteases,
Cannabis, Viral
Class B: Secretin-like Family
Glucagon, Calcitonin, parathyroid hormone, secretin
Class C: Metabotropic glutamate and Chemosensor Family
mGluR 1-7, Calcium sensors, GABA-B
Class D: Fungal pheromone Family
Class E: c-AMP receptor ( Dictyostelium) Family
Class F: Frizzled/Smoothened family
Putative families:
Ocular albinism proteins, Drosophila odorant receptors, Plant Mlo receptors,Nematode
chemoreceptors, Vomeronasal receptors
Putative/ unclassified orphans
Structure of GPCRs
C-tail
• Only one structure
known, rhodopsin
• rhodopsin serves
as model for other
pharmacologically
important GPCR
• Some GPCR share
less than 10%
sequence identity
C-II
C-I
C-III
VIII
VI
VII
V
IV
II
I
III
Disulfide Bond
Cys110-Cys187
4
3
E-II
E-I
2
E-III
1
N-tail
Previous Template:
Bacteriorhodopsin
Mammalian Rhodopsin = magenta
Halorhodopsin = gray
Sensory Rhodopsin = blue
Improvements
• Initial homology model can be refined by
molecular dynamics simulation
• including lipid bilayer in the modeling
process has been shown to give better
models
Examples for Homology Modeling
of GPCRs
•
•
•
•
b2-adrenergic receptor
angiotensin II receptor type I (AT1)
purinergic GPCRs
monocyte chemoattractant-1 (MCP-1) receptor, CCR2
In all cases, models were able to explain site-directed mutagenesis
data and could be used to dock ligands.
However, ligand-based lead finding (often using the natural ligand as a
starting point) is still most widely used for GPCR. Structure-activity
relationships (SAR) are derived from them and the resulting
pharmacophore models can be used for virtual screening. Need
more structures!
First global network on structural genomics on GPCRs (MePNet):
http://www.mepnet.org
Status: http://www.mepnet.org/index.php?rub=mepnet_1101_0204
Summary GPCR Case Study
• Being able to model proteins with low
sequence homology is essential to exploit
structural information that is hard to get
(membrane proteins) but where the impact
is very high (>40% of R&D portfolios in
companies)
Take home messages
• Structural and functional effects of small changes in
sequences
• Conservation of structure despite large differences in
sequences
• Prediction of structural and functional effects using
computational pharmacology to understand disease
mechanisms and drug action with the goal of identifying
targets and designing drugs against them
– Example: Specific Structure of COX and of GPCR
– Current hot topics: Complex interactions of proteins
within their environment, differences between
individuals, systems biology
• Even with lots of structural information available,
prediction of ligand binding affinities is challenging
• Determination of new structures, especially membrane
proteins, is a bottle-neck