* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Biosynthesis of proteins on ribosomes GENETIC
Polyadenylation wikipedia , lookup
Magnesium transporter wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Protein moonlighting wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Molecular evolution wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Western blot wikipedia , lookup
List of types of proteins wikipedia , lookup
Homology modeling wikipedia , lookup
Non-coding RNA wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Peptide synthesis wikipedia , lookup
Gene expression wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Point mutation wikipedia , lookup
Protein adsorption wikipedia , lookup
Messenger RNA wikipedia , lookup
Bottromycin wikipedia , lookup
Protein structure prediction wikipedia , lookup
Biochemistry wikipedia , lookup
Epitranscriptome wikipedia , lookup
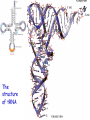
Transfer RNA wikipedia , lookup
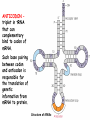
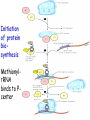
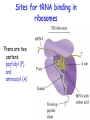
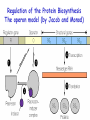
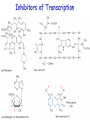
Biosynthesis of proteins on ribosomes GENETIC CODE sequence of mononucleotides in mRNA that specifies the sequence of amino acids in peptide chain CODON – mRNA triplet base sequence responsible for 1 amino acid PROPERTIES OF GENETIC CODE 1. Unambiguous. In any organism each codon corresponds to only one amino acid. 2. Code is degenerate. There are multiple codons for most amino acids. 3. Universal. Codons are the same for all organism. 4. Without punctuation. There are no punctuations between trinucleotides. 5. Nonoverlapping. Codons do not overlap each other. ANTICODON – triplet in tRNA that can complementary bind to codon of mRNA. Such base pairing between codon and anticodon is responsible for the translation of genetic information from mRNA to protein. Structure of tRNAs STAGES OF TRANSLATION • 1. Recognition • 2. Initiation • 3. Elongation • 4. Termination RECOGNITION O R1 CH COOH + H O O P O P OH NH2 O O OH P O Аденозин OH O R1 CH CO O P O Аденозин + H4P2O7 OH NH2 Aminoacyladenilate Aminoacyl-tRNA-synthetase Aminoacyladenilate + tRNA aminoacyl-tRNA + AMP Activation of amino acids Each amino acid has a specific tRNA There is specific aminoacyl-tRNA-synthetase for each AA The structure of tRNA Initiation of Translation • The translation complex is assembled at the beginning of the mRNA coding sequence • Complex consists of: -Ribosomal subunits -mRNA template to be translated -Initiator tRNA molecule -Protein initiation factors Initiator tRNA • First codon translated is usually AUG • The initiator tRNA recognizes initiation codons -Bacteria: N-formylmethionyl-tRNA -Eukaryotes: methionyl-tRNA Initiation of protein biosynthesis MethionylтRNA binds to Pcenter Sites for tRNA binding in ribosomes There are two centers: peptidyl (P) and aminoacyl (А) Elongation 1) Positioning of the next aminoacyl-tRNA in the A site 2) Formation of the peptide bound (enzyme – peptidyl transferase) between methionine and AA in Acentre. The residue of methionine is transferred on the amino group of another AA 3) Translocation – shift of ribosome by one codon. Methionyl-tRNA is released from P-centre. DipeptidyltRNA moves from A-centre to P-centre. Termination of Translation • Ribosome comes to terminal codon UGA, UAG or UAA • No tRNA molecules recognize these codons and protein synthesis stalls • Protein termination factors F-1, RF-2, RF-3 split off synthesized polypeptide from the last tRNA • Ribosomal complex dissociates Termination of Translation POSTTRANSLATIONAL MODIFICATION 1) Preparing of proteins for different functions 2) Direction of proteins to different locations (targeting) 1. Removing of methionine (formylmethionine) 2. Formation of disulfide and other bonds (secondary, tertiary structures) 3. Proteolytic cleavage 4. Modification of amino acid residues: - Hydroxylation - Glycosilation - Phosphorilation 5. Joining of prosthetic groups or cofactors 6. Formation of the quaternary structure Regulation of the Protein Biosynthesis The operon model (by Jacob and Monod) Inhibitors of Transcription Antibiotics inhibiting protein synthesis