* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download second of Chapter 10: RNA processing
History of genetic engineering wikipedia , lookup
Transposable element wikipedia , lookup
Microevolution wikipedia , lookup
Transcription factor wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Frameshift mutation wikipedia , lookup
Metagenomics wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Human genome wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Point mutation wikipedia , lookup
Expanded genetic code wikipedia , lookup
Transfer RNA wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Non-coding DNA wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
RNA interference wikipedia , lookup
Alternative splicing wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Genetic code wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Nucleic acid tertiary structure wikipedia , lookup
RNA silencing wikipedia , lookup
History of RNA biology wikipedia , lookup
Non-coding RNA wikipedia , lookup
RNA-binding protein wikipedia , lookup
Messenger RNA wikipedia , lookup
Polyadenylation wikipedia , lookup
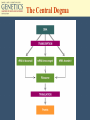
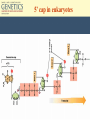
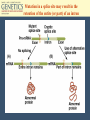
Chapter 10 Transcription RNA processing Translation Jones and Bartlett Publishers © 2005 Terms (mRNA) • • • • primary transcript coding sequence (open reading frame, ORF) prokaryotic mRNA = primary transcript Eukaryotic transcripts are converted into mRNA through RNA processing: – Modification of the 5’ end – Extension of 3’ end – Excision of untranslated embedded sequences. The Central Dogma Polycistronic messages • In bacteria and eukaryotic organelles, sometimes more than one polypeptide is encoded in a single message. • These are called polycistronic mRNAs. Polycistronic mRNAs Eukaryotic Transcription • In eukaryotes, three different polymerases are used. • No polycistronic mRNAs are produced. (All eukarytoic mRNAs are monocistronic.) • The mRNA molecules are chemically modified in eukaryotes. Eukaryotic RNA Polymerases Enzyme Cellular location RNA polymerase I Nucleolus Product rRNA RNA polymerase II Nucleus mRNA snRNA RNA polymerase III Nucleus tRNA, 5S RNA, snRNA Promoters for eukaryotic genes • There is more variability in promoters and termination sequences. • TATA box : 5’-TATAAA-3’, -25 bp, with GC regions near it. • CAAT box: GCCAATCT, -80 bp • GC box: GGGCGC, multiple copies. Promoters in eukaryotes Modification of primary transcripts • Three major alterations to mRNA: – Modified guanosine cap at 5’ end – 3’ end gets a poly(A) tail. – pre-mRNA is processed to remove specific internal sequences by splicing them out. 5’ cap in eukaryotes Polyadenylation • The poly(A) tail is added by poly(A) polymerase. • About 200 adenosines are added to the 3’ end of the primary transcript. • Poly(A) tails help geneticists isolate mRNAs from cells. RNA Splicing • In the 1960s, it was observed that RNA molecules were larger than predicted, based on protein structure. • In 1977, internal, non-coding sequences were discovered. • These internal, non-coding sequences are called introns. Spliceosomes and the GU-AG rule • Intron-Exon junctions have a conserved sequence that is recognized by an enzyme complex (spliceosome). • The sequence is GU-AG, and is almost universal. • A spliceosome has nucleic acids, 8-10 proteins, and snRNAs. RNA processing Transcription and RNA processing are coupled processes. Molecular details of the splicing of an intron Introns begin with a GU and end with an AG. There is also an internal A nucleotide where the branch junction is formed. The spliced intron is known as a lariat (with a loop and tail). Splicing of introns is mediated by small nuclear ribonucleoprotein particles (snRNPs) In this diagram, only the RNA component of the snRNPs is shown. Base pairing between snRNA and the intron is involved in the splicing reaction. DNA-RNA hybridization showing the parts of DNA that are missing from the mature mRNA and the part of the mRNA (polyA tail) missing in the DNA Mutation in a splice site may result in the retention of the entire (or part) of an intron Exon-shuffle model • Introns may play a role in gene evolution. • In some proteins, each exon has its own independent folding characteristics. • Folding domains (=exons) can be grouped together to give new proteins with new functions. • This is called the exon-shuffle model. • Not all genes have domain boundaries that correlate to exons. Transcription occurs in the nucleus, as does RNA processing. Export to the cytoplasm happens before translation occurs. Translation of mRNA into amino acid sequences • The genetic code consists of three bases. • A codon is the ‘code word’ for each amino acid. • 4 x 4 x 4 =64, but only 20 amino acids are used in proteins, so the genetic code is said to be degenerate.