* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download PROTEIN STRUCTURE SIMILARITY CALCULATION AND VISUALIZATION
Cell-penetrating peptide wikipedia , lookup
Expanded genetic code wikipedia , lookup
Genetic code wikipedia , lookup
Gene expression wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Magnesium transporter wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Biochemistry wikipedia , lookup
Interactome wikipedia , lookup
Protein moonlighting wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein folding wikipedia , lookup
Circular dichroism wikipedia , lookup
Protein domain wikipedia , lookup
Western blot wikipedia , lookup
Protein adsorption wikipedia , lookup
Homology modeling wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Structural alignment wikipedia , lookup
PROTEIN STRUCTURE
SIMILARITY CALCULATION
AND VISUALIZATION
CMPS 561-FALL 2014
SUMI SINGH
SXS5729
Protein Structure
RPDFCLEPPYAGACRARIIRYFYNAKAGLCQ
Primary Structure
Sequence of Amino Acids.
Not enough for functional prediction.
Tertiary Structure
(3D Structure)
Formed by 3D folding
pattern of the protein. It
makes protein functional.
2
Comparing protein 3D structuresget functional insight
Structure of 1QLQ
Structure of 4HHB
Compare structures of two
DIFFERENT proteins
3
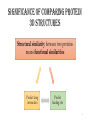
Significance of comparing protein
3D structures
Structural similarity between two proteins
means functional similarities
Predict drug
interaction
Predict
binding site
4
Structural elements represented
by quintuple of features
{𝐿𝑎𝑏𝑒𝑙1 , 𝐿𝑎𝑏𝑒𝑙2 , 𝐿𝑎𝑏𝑒𝑙3 , 𝜃, 𝐷}
Labels represent
Primary
Structure (amino
acids sequence)
Tertiary/ 3D
structure
Theta represents
orientation
Length
represents
size/scale
5
Structural alphabet (key)
generation
Generate all possible
triples of amino acids 𝐶3𝑛
Assign labels to
amino acids in triple
Perform rule based
label arrangement
Label3
{𝐿𝑎𝑏𝑒𝑙1 , 𝐿𝑎𝑏𝑒𝑙2 , 𝐿𝑎𝑏𝑒𝑙3 , 𝜃, 𝐷}
Quintuple
Mapping from structure
space into unique key
(integer space)
d23
d13
Label1
d12
Calculate Angle
and Length
Label2
Representative Length
(D)
6
Output of the key generation
system
For every protein millions of keys are generated each
representing some special feature.
The protein structure is represented and stores as unique
KEY-COUNT pair.
Learning goals
Familiarizing with complex research problem and the process of
solving it including reading and understanding published
research papers and using them in problem solving.
Parallel implementation of algorithm(s) and
demonstrate the speedup from serial to parallel.
Visualizing the output.
Task Outline
Calculate pairwise similarity between two
proteins implemented in PARALLEL (moduleA)
Structure of 1QLQ
TSR Key-Count Set representing 1QLQ
Structure of 4HHB
TSR Keys-Count Set representing 4HHB
Similarity Computation
Jaccard Coefficient that allows (unique or count={0,1}) set as its arguments
Jaccard-Tanimoto Coefficient that allows multi-sets (count>1) as its arguments
11
Input to moduleA
All input files will be given as key-count pairs that will be the input to the
system.
Keys are integers representing the unique structural feature.
All keys for a given protein will have corresponding count >=1.
There may be some keys that present in one protein while absent in other as they
represent unique features.
Output from moduleA
You will be given a set of proteins and you have to calculate all by all
pairwise similarity between them.
Display/write the pairwise similarity between each protein file as lower triangular
matrix for comparison purpose
Input to moduleB or visualization
module and the output
The all by all pairwise similarity calculated in moduleA will be used as
input to moduleB.
Output should be connectivity graph (as shown in next slide) between
all proteins.
Each edge must display the similarity value.
Preferred output will be each edge length weighted as similarity value
between the two connecting proteins.
Construct structural similarity graph
(moduleB)
Method for finding the global structural connectivity between proteins that
contain a specific domain of interest.
84
%
1A06
80%
84%
1AD5
85%
1FMK
85%
74%
74%
1FGK
74%
1ERK
83%
75%
75%
1CKI
15
Final system
Should integrate moduleA and moduleB.
If given a set of proteins should be able to find all by all
similarity between them, display the lower triangular
similarity matrix.
Construct similarity graph.
What do you get from me?
1. Training protein structure (key-count) file with their precalcuated
similarity values, both Jaccard and Jaccard Tanimoto
-- around 50 proteins
-- you can use these to evaluate your system
2. Test set (50 proteins), only key-count pairs and no similarity values.
3. All the files will be text files.
4. Time taken by me to calculate the all by all similarity on the test and
training set using an optimized serial algorithm for comparison
with your parallel implementation.
You can use Hadoop-mapreduce for moduleA.
Visualization can be done on GEPHI
http://gephi.github.io/
Information on Jaccard and Jaccard-Tanimoto can be found in the following
paper:
http://csis.pace.edu/ctappert/dps/d861-12/session4-p2.pdf
Lower triangular matrix:
http://en.wikipedia.org/wiki/Triangular_matrix