* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download video slide - Saginaw Valley State University
Non-coding DNA wikipedia , lookup
RNA interference wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
List of types of proteins wikipedia , lookup
Community fingerprinting wikipedia , lookup
Promoter (genetics) wikipedia , lookup
RNA silencing wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Gene regulatory network wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Polyadenylation wikipedia , lookup
Molecular evolution wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Bottromycin wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Non-coding RNA wikipedia , lookup
Biochemistry wikipedia , lookup
Messenger RNA wikipedia , lookup
Gene expression wikipedia , lookup
Genetic code wikipedia , lookup
Transfer RNA wikipedia , lookup
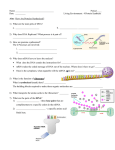
Chapter 17: From Gene to Protein Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings Figure 17.1 A ribosome, part of the protein synthesis machinery Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings Figure 17.2 Do individual genes specify different enzymes in arginine biosynthesis? EXPERIMENT Working with the mold Neurospora crassa, George Beadle and Edward Tatum had isolated mutants requiring arginine in their growth medium and had shown genetically that these mutants fell into three classes, each defective in a different gene. From other considerations, they suspected that the metabolic pathway of arginine biosynthesis included the precursors ornithine and citrulline. Their most famous experiment, shown here, tested both their one geneone enzyme hypothesis and their postulated arginine pathway. In this experiment, they grew their three classes of mutants under the four different conditions shown in the Results section below. RESULTS The wild-type strain required only the minimal medium for growth. The three classes of mutants had different growth requirements Wild type Minimal medium (MM) (control) MM + Ornithine MM + Citrulline MM + Arginine (control) Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings Class I Mutants Class II Mutants Class III Mutants CONCLUSION From the growth patterns of the mutants, Beadle and Tatum deduced that each mutant was unable to carry out one step in the pathway for synthesizing arginine, presumably because it lacked the necessary enzyme. Because each of their mutants was mutated in a single gene, they concluded that each mutated gene must normally dictate the production of one enzyme. Their results supported the one gene–one enzyme hypothesis and also confirmed the arginine pathway. (Notice that a mutant can grow only if supplied with a compound made after the defective step.) Class I Class II Class III Mutants Mutants Mutants (mutation (mutation (mutation in gene A) Wild type in gene B) in gene C) Precursor Gene A Enzyme A Ornithine Gene B Enzyme B Citrulline Gene C Enzyme C Arginine Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings Precursor Precursor Precursor A A A Ornithine Ornithine Ornithine B B B Citrulline Citrulline Citrulline C C C Arginine Arginine Arginine Figure 17.4 The triplet code DNA molecule Gene 2 Gene 1 Gene 3 DNA strand (template) 5 3 A C C A A A C C G A G T U G G U U U G G C U C A TRANSCRIPTION mRNA 5 Codon TRANSLATION Protein Trp Amino acid Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings Phe Gly Ser 3 Figure 17.5 The dictionary of the genetic code Second mRNA base C UUU UUC U UUA First mRNA base (5 end) A UAU UCU Phe UCC UCA UAC Ser U UGU Tyr UGC Cys C UCG CUU CCU CAU CUC CCC CAC CUA Leu Leu CCA Pro CAA CUG CCG CAG AUU ACU AAU ACC AAC AUC lle AUA ACA Thr AAG GUU GCU GAU GUC GCC GAC GUA GUG Val GCA Ala GCG Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings His Gln Asn AAA AUGMet or ACG start G G UAA Stop UGA Stop A UAG Stop UGG Trp G UUG C A Lys CGC CGA Arg Asp CGG G AGU U AGC Ser C AGA A AGG Arg G GGC GGA Glu C A U GGU GAA GAG U CGU GGG Gly C A G Third mRNA base (3 end) U Figure 17.6 A tobacco plant expressing a firefly gene Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings Non-template strand of DNA Elongation RNA nucleotides RNA polymerase A T C C A A 3 3 end U 5 A E G C A T A G G T T Direction of transcription (“downstream) 5 Newly made RNA Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings Template strand of DNA Figure 17.13 Translation: the basic concept TRANSCRIPTION DNA mRNA Ribosome TRANSLATION Polypeptide Amino acids Polypeptide Ribosome tRNA with amino acid attached Gly tRNA Anticodon A A A U G G U U U G G C Codons 5 mRNA Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings 3 Figure 17.14 The structure of transfer RNA (tRNA) 3 A Amino acid C attachment site C A 5 C G G C C G U G U A A U A U U C * G U AG * CACA A CUC * G * U G U G G * C CGAG C * * AGG U * * GA G C Hydrogen G C bonds U A * G * A A C * U A A G Anticodon (a) Two-dimensional structure. The four base-paired regions and three loops are characteristic of all tRNAs, as is the base sequence of the amino acid attachment site at the 3 end. The anticodon triplet is unique to each tRNA type. (The asterisks mark bases that have been chemically modified, a characteristic of tRNA.) Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings 5 3 Amino acid attachment site Hydrogen bonds A 3 Anticodon (b) Three-dimensional structure Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings A G Anticodon 5 (c) Symbol used in this book Figure 17.15 An aminoacyl-tRNA synthetase joins a specific amino acid to a tRNA Amino acid Aminoacyl-tRNA synthetase (enzyme) 1 Active site binds the amino acid and ATP. P P P Adenosine ATP 2 ATP loses two P groups and joins amino acid as AMP. P Adenosine Pyrophosphate Pi Phosphates P Pi Pi tRNA 3 Appropriate tRNA covalently Bonds to amino Acid, displacing AMP. P Adenosine AMP 4 Activated amino acid is released by the enzyme. Aminoacyl tRNA (an “activated amino acid”) Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings Figure 17.16 The anatomy of a functioning ribosome TRANSCRIPTION DNA mRNA Ribosome TRANSLATION Polypeptide Exit tunnel Growing polypeptide tRNA molecules Large subunit E P A Small subunit 5 mRNA 3 (a) Computer model of functioning ribosome. This is a model of a bacterial ribosome, showing its overall shape. The eukaryotic ribosome is roughly similar. A ribosomal subunit is an aggregate of ribosomal RNA molecules and proteins. Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings Figure 17.23 The molecular basis of sickle-cell disease: a point mutation Wild-type hemoglobin DNA 3 Mutant hemoglobin DNA 5 C T T In the DNA, the mutant template strand has an A where the wild-type template has a T. A The mutant mRNA has a U instead of an A in one codon. 3 5 T C mRNA A mRNA G A A 5 G 3 U 5 3 Normal hemoglobin Sickle-cell hemoglobin Glu Val Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings The mutant (sickle-cell) hemoglobin has a valine (Val) instead of a glutamic acid (Glu). Figure 17.26 A summary of transcription and translation in a eukaryotic cell DNA TRANSCRIPTION 1 RNA is transcribed from a DNA template. 3 5 RNA transcript RNA polymerase RNA PROCESSING Exon 2 In eukaryotes, the RNA transcript (premRNA) is spliced and modified to produce mRNA, which moves from the nucleus to the cytoplasm. RNA transcript (pre-mRNA) Intron Aminoacyl-tRNA synthetase NUCLEUS Amino acid tRNA FORMATION OF INITIATION COMPLEX CYTOPLASM 3 After leaving the nucleus, mRNA attaches to the ribosome. mRNA AMINO ACID ACTIVATION 4 Each amino acid attaches to its proper tRNA with the help of a specific enzyme and ATP. Growing polypeptide Activated amino acid Ribosomal subunits 5 TRANSLATION A succession of tRNAs add their amino acids to the polypeptide chain Anticodon as the mRNA is moved through the ribosome one codon at a time. (When completed, the polypeptide is released from the ribosome.) 5 E A AAA UG GU U U A U G Codon Ribosome Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings