* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Bio1A Unit 1-3 The Cell Notes File
Survey
Document related concepts
Protein (nutrient) wikipedia , lookup
Cytoplasmic streaming wikipedia , lookup
SNARE (protein) wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Magnesium transporter wikipedia , lookup
Cytokinesis wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein moonlighting wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Extracellular matrix wikipedia , lookup
Cell nucleus wikipedia , lookup
Cell membrane wikipedia , lookup
Signal transduction wikipedia , lookup
Western blot wikipedia , lookup
Proteolysis wikipedia , lookup
Transcript
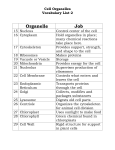
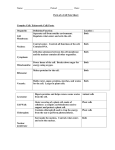
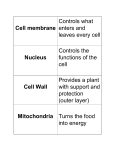
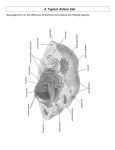
The Cell Organelles & Their Functions Nucleus Ribosomes Lysosome Peroxisome Mitochondria Plastids - Chloroplasts Cytoskeleton Extracellular Matrix Protein Trafficking Membranes Membrane fluidity Membrane Proteins The Cell Major Features 1. Membrane – compartmentalization • Separates inside and outside • But there is flow in and out Material Communication 2. Cytosol – semifluid substance 3. Genetic Material: i.e.- DNA • Directs cells activity • Allows reproduction 4. Ribosomes – Protein production Prokaryotic vs no nucleus Single circular DNA / chromosome No membrane bound organelles Eukaryotic Eukaryotes – nucleus contains DNA Linear pieces of DNA / chromosomes Membrane bound organelles Organelles Nucleus •Contains most of the Cell’s DNA (as chromatin) •Nuclear pores for transport Pore proteins determine what goes in and out •Continuous with ER Ribosome – NOT (technically) an organelle Function - Protein Translation : RNA Protein Location - Free floating in cytoplasm or bound to endoplasmic reticulum Stucture - 2 multi-complex proteins composed of protein and rRNA named according to relative size (small and large). Endoplasmic Reticulum (ER) Smooth ER – Lipid biosynthesis – ex: steroids, Fats Carbohydrate metabolism – breakdown of polysaccharides Detoxification of drugs NOT protein synthesis – no ribosomes Rough ER – rough due to presence of ribosomes 1. Protein synthesis 2. Protein modifications 3. Glycosylation Carbohydrates (CH2O groups) are attached to some proteins 4. Transferred to Golgi through transport vesicles (no glycosylation = stays in ER) Golgi apparatus - Protein Transport / Protein Modification • • • Phosphorylation lysosome Glycosylation deglycosylation Modification determines final destination Lysosome - Digestive organelle • • pH 4-5 (required for enzyme function) Lysosomal enzymes can hydrolyze proteins, fats, polysaccharides, and nucleic acids Some types of cell can engulf another cell by phagocytosis; this forms a food vacuole A lysosome fuses with the food vacuole and digests the molecules Lysosomes also use enzymes to recycle the cell’s own organelles and macromolecules, a process called autophagy Peroxisomes: Oxidation • Peroxisomes are specialized metabolic compartments bounded by a single membrane • Peroxisomes produce hydrogen peroxide and convert it to water • Oxygen is used to break down different types of molecules Mitochondria and chloroplasts change energy from one form to another – Are not part of the endomembrane system – Have a double membrane – Contain their own circular DNA & ribosomes (small) ~20% of protein made inside • Mitochondria (plants and animals) are the sites of cellular respiration, a metabolic process that generates ATP • Chloroplasts (plants and algae) are the sites of photosynthesis - Part of a subcategory of plant/algae organelle called plastids Vacuole – plant •Large storage vesicle cytoskeleton • Fiber network throughout the cytoplasm • Organizes cell’s structures and activities, anchoring many organelles – support the cell and maintain its shape – interacts with motor proteins to produce motility I. Microtubules - Shaping the cell • Thickest • Guiding movement of organelles • Separating chromosomes during cell division I. Microfilaments – propulsion • control the beating of cilia and flagella, locomotor appendages of some cells • Cilia and flagella differ in their beating patterns II. Intermediate filaments: Actin – cellular motility • Thinnest • built as a twisted double chain of actin subunits • structural role - to bear tension, resisting pulling forces within the cell The Extracellular Matrix (ECM) • Animal cells lack cell walls but are covered by an elaborate extracellular matrix (ECM) • made of glycoproteins such as collagen (fibrous), proteoglycans, and fibronectin • ECM proteins bind and anchor, and transmit messages Protein trafficking Post-Translational Import - Proteins made on free ribosomes Final Destinations cytosol, mitochondria & chloroplast, peroxisomes, nucleus (usually) Requires a transport protein which targets: N-terminal (usually) targeting signal sequence of peptide Some have names. Examples: • Nucleus – Nuclear Localization Signal (NLS) Importin – protein that transports NLS tagged proteins • Peroxisomal Targeting Signal CO-Translational Import - Proteins made on bound ribosomes / Rough ER Final Destinations ER, Golgi, lysosome, nuclear membrane, membrane, extracellular Targets membranes called endomembrane system Amino acid sequences are targets for • directing translating ribosome to bind to ER rough ER • Are targets for protein modifications in ER or golgi Protein modification In ER Glycosylation: Carbohydrates (CH2O groups) are attached to some proteins Glycosylation causes transport to Golgi through transport vesicles (no glycosylation = stays in ER) In golgi Further modification determines final location Plasma Membrane • the boundary that separates the living cell from its surroundings • exhibits selective permeability, allowing some substances to cross it more easily than others • Basis for permeability is the hydrophobic nature of the tails of phospholipids - Lipid bilayer - Naturally forms do to hydrophillic interaction of the heads which must face both outward and inward to the water environment both outside and inside the cell Fluid Mosaic Model • The lack of even weak hydrogen bonding results in a fluid membrane (see video) • The plasma membrane is a collage spotted with many different proteins (mosaic) ~45% protein 45% phospholipid 10% other • Proteins determine most of the cell’s function Cholesterol stabilizes membranes @ high temp – slows fluidity @ low temp – makes more fluid Membrane Proteins Characteristics • Peripheral – bound (loosely) to surface • Integral = transmembrane proteins (tightly bound) hydrophobic regions - stretches of nonpolar amino acids, often coiled into alpha helices