* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Fluorescent dye, SYBR Green, is incorporated into PCR reaction
Zinc finger nuclease wikipedia , lookup
Epigenetics of human development wikipedia , lookup
DNA polymerase wikipedia , lookup
Pathogenomics wikipedia , lookup
Transposable element wikipedia , lookup
Oncogenomics wikipedia , lookup
DNA sequencing wikipedia , lookup
DNA profiling wikipedia , lookup
X-inactivation wikipedia , lookup
Mitochondrial DNA wikipedia , lookup
DNA damage theory of aging wikipedia , lookup
Whole genome sequencing wikipedia , lookup
SNP genotyping wikipedia , lookup
Primary transcript wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Point mutation wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Genetic engineering wikipedia , lookup
DNA vaccination wikipedia , lookup
Minimal genome wikipedia , lookup
United Kingdom National DNA Database wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Human genome wikipedia , lookup
Comparative genomic hybridization wikipedia , lookup
Genome (book) wikipedia , lookup
Genealogical DNA test wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Epigenomics wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Deoxyribozyme wikipedia , lookup
DNA supercoil wikipedia , lookup
Microsatellite wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Metagenomics wikipedia , lookup
Genome evolution wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Non-coding DNA wikipedia , lookup
Designer baby wikipedia , lookup
Molecular cloning wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Microevolution wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Genome editing wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Helitron (biology) wikipedia , lookup
History of genetic engineering wikipedia , lookup
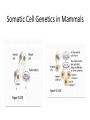
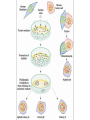
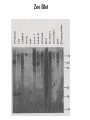
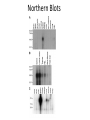
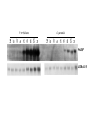
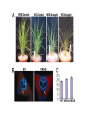
PHYSICAL MAPPING AND POSITIONAL CLONING • Linkage mapping – Flanking markers identified – 1cM, for example • Probably ~ 1 MB or more in humans • Need very many families to get closer than this in human, or very large populations What to Do Next? • Identify genes in this region • Then determine what is the gene of interest What to do next? • In 1990, clone region around markers, make physical map(s), look for genes experimentally • In 2000, use better physical maps, at least in some organisms • In 2010, use sequencing, bioinformatic knowledge, experimental proof still necessary Physical Mapping A physical map is primarily based on the locations of landmarks along a DNA molecule and units of distance are expressed in base pairs. Low Resolution Physical Mapping • Cytogenetic map • In situ hybridization Chromosome Sorting • Flow cytometry • Used in library construction • Also for chromosome paints Somatic Cell Genetics in Mammals Radiation Hybrid Mapping Top-down mapping Bottom-up Mapping (Contig Maps, Mapping With Ordered Clones) Cloning Vectors • • • • • • Plasmid Phagemid Cosmid, fosmid YACs BACs PACs Application Of BAC library Chromosome walking & contig assembly Whole genome sequencing Fluorescent in situ hybridization BAC library Fine mapping for interest gene Positional cloning Construction of integrated genetic and physical map Maximum pyrosequencing read lengths currently are 300-500 nt. Commercial applications: 454 Life Sciences Genome Sequencer FLX Generate 400 million nt in 10 hours $5-7K USD per run $1M for mammalian genome Zoo Blot CpG islands Northern Blots 1d 12h 8h 4h 2h 1h 30’ 15’ NA 1d 12h 8h 4h 2h 1h 30’ 15’ NA P. trifoliata C. paradisi PtCBF CORc115 Real-time Quantitative PCR: Measures the abundance of DNA as it is amplified. Useful for quantitatively measuring the levels of mRNA in a sample. Uses reverse transcriptase to generate cDNA for the template. Can also be used to quantitatively estimate fraction of DNA from various organisms in a heterogenous sample (e.g, can be used to measure abundance of different microbes in soil sample). Fluorescent dye, SYBR Green, is incorporated into PCR reaction. SYBR Green fluoresces strongly when bound to DNA, but emits little fluorescence when not bound to DNA. SYBR Green fluorescence is proportional to the amount of DNA amplified, detected with a laser or other device. Experimental samples are compared to control sample with known concentration of cDNA. Fig. 10.9 SYBR Green binds to double-stranded DNA and fluoresces Sequence Conservation • Homology searches Array Technology Gene Complementation • • • • • Mutants Overexpression “Knockouts” RNAi Reporter assays