* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Chapter 3 (part 2) – Protein Function
Lipid signaling wikipedia , lookup
Biochemical cascade wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Multi-state modeling of biomolecules wikipedia , lookup
Paracrine signalling wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Biosynthesis wikipedia , lookup
Protein purification wikipedia , lookup
Western blot wikipedia , lookup
Interactome wikipedia , lookup
Clinical neurochemistry wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Biochemistry wikipedia , lookup
Drug design wikipedia , lookup
Signal transduction wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Metalloprotein wikipedia , lookup
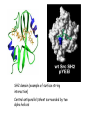
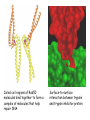
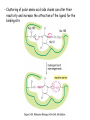
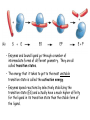
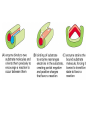
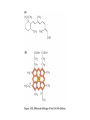
Chapter 3 (part 2) – Protein Function Test Your Knowledge • (True/False) All proteins bind to other molecules. Explain. • What sort chemical interactions create the “binding” between the ligand and its protein partner? Name at least two. • (True/False) A ligand binding site can be kept dry from surrounding water molecules. Explain. • Explain two different ways (structural) that proteins typically bind to other proteins. • What is Km? What does having a low Km say about the binding between a protein and its ligand? • What are some of the major functions of proteins in a cell? Name at least 3 “categories”. Protein-Protein Binding Three major mechanisms: • “surface string” interaction – • “helix-helix” or “coiled coil” interaction – • “surface-surface” interaction – SH2 domain (example of surface string interaction) Central antiparallel bsheet surrounded by two alpha helices Coiled coil regions of Rad50 molecules bind together to form a complex of molecules that help repair DNA Surface-to-surface interaction between trypsin and trypsin inhibitor protein Protein-Ligand Interactions Protein Binding Sites –Sterically and chemically “fit” the ligand; Examples are antibodies and enzymes • Water is typically excluded from the binding site, but is important to overall structure. Why? • Clustering of polar amino acid side chains can alter their reactivity and increase the attraction of the ligand for the binding site cAMP bound to a binding site on a protein • Enzymes and bound ligand go through a number of intermediate forms of different geometry. They are all called transition states. • The energy that it takes to get to the most unstable transition state is called the activation energy. • Enzymes speed reactions by selectively stabilizing the transition state (ES) and actually have a much higher affinity for the ligand in its transition state than the stable form of the ligand. • Enzymes not only bind to the substrate, but the nature of the amino acids and their side chains within the binding site alter chemical bonds in the substrate. • Many enzymes can perform both acid and base catalysis because of the different side chains in their binding site. This allows increased speed of catalysis. Many Enzymes Require Tightly Bound Small Molecules to Function • Enzymes sometimes form complexes that also improve efficiency of reactions that may otherwise be diffusion-limited. •Examples – pyruvate dehydrogenase, tryptophan synthase, aminoacyl t-RNA synthetase