* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Lecture 19 - University of Wisconsin–Madison
Survey
Document related concepts
Proteolysis wikipedia , lookup
NADH:ubiquinone oxidoreductase (H+-translocating) wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Restriction enzyme wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Enzyme inhibitor wikipedia , lookup
Ligand binding assay wikipedia , lookup
Biosynthesis wikipedia , lookup
Photosynthetic reaction centre wikipedia , lookup
Structural alignment wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Catalytic triad wikipedia , lookup
Metalloprotein wikipedia , lookup
Transcript
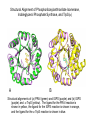
Chemical mechanism is dominant • Nature selects the protein for divergent evolution from a pool of enzymes whose mechanism provide a partial mechanism, or provide the means to stabilize an energetically unfavorable intermediate or transition state. • The original enzyme might have acquired a low level of the new activity through adventitious mutations. Subsequently the new enzyme would evolve (selective pressure) to increase the proficiency of the new reaction at the loss of its original. • The old and new enzymes would be homologous and belong to a mechanistically diverse superfamily. • Examples of this type of evolution are seen in the enolase, amidohydrolase/phosphotriesterase, and crotonase superfamilies. Structural Alignment of Phosphoribosylanthranilate Isomerase, Indoleglycerol Phosphate Synthase, and TrpS(a) A B Structural alignments of (a) PRAI (green) and IGPS (purple) and (b) IGPS (purple) and a-TrpS (yellow). The ligand for the PRAI reaction is shown in yellow, the ligand for the IGPS reaction is shown in orange, and the ligand for the a-TrpS reaction is shown in blue. Superposition of OMPDC and KGPDC Structural alignment of 3-Keto-L-gulonate 6-phosphate decarboxylase (KGPDC) (purple) with bound L-gulonate 6-phosphate (yellow) and OMPDC (green) with bound UMP (orange). Although they share limited sequence identity, both enzymes adopt a conserved (a)8 barrel fold. The quaternary relationship between the two individual subunits in the dimer is highly conserved. Location of Catalytic groups in the Super Family Strand 1 2 Enolase 3 4 5 6 7 8 D E DE K (H) (K) H E MR KxK D EEP E D MLE KxK D EQP DE K (E) Base blue, acid red, and Mg2+ ligands green. A striking feature of this super family is the conserved location of the catalytic groups. This supports the hypothesis that these enzymes arose by divergent evolution, since there is no reason that the functional groups should lie on the same strand. The relative orientation (order) of the groups around the barrel is required to provide adequate coordination of the metal and to place the catalytic bases on opposite sides of the substrate. There is no strict structural requirement that the metal ligands lie on the third, fourth, and fifth strands. OMPDC and KGPDC Use the Same Active Site Residues for Different Purposes The active site resides in OMPDC and KGPDC are remarkably similar, yet they serve different purposes in their reaction schemes. Conclusions • It seems likely that in one case the (/a)8 barrel arose via gene duplication of a smaller folding domain. • Did all (/a)8 barrels arise with this mechanism? Probably not. • Structures provide insight into enzyme evolution, but not always answers!