* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Biosynthesis and degradation of proteins
Histone acetylation and deacetylation wikipedia , lookup
Biochemistry wikipedia , lookup
Gene expression wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Index of biochemistry articles wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Magnesium transporter wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Proteases in angiogenesis wikipedia , lookup
Catalytic triad wikipedia , lookup
Signal transduction wikipedia , lookup
Protein folding wikipedia , lookup
Interactome wikipedia , lookup
Protein moonlighting wikipedia , lookup
Paracrine signalling wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Western blot wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein structure prediction wikipedia , lookup
Protein adsorption wikipedia , lookup
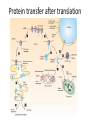
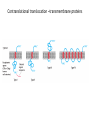
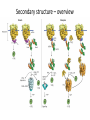
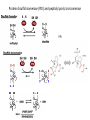
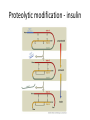
Biosynthesis and degradation of proteins Bruno Sopko Content • Proteosynthesis • Post-translation processing of proteins • Protein degradation Proteosynthesis • • • • Aminoacyl-tRNA formation Iniciation Elongation Termination Aminoacyl-tRNA formation Amino acid + ATP ↔ Aminoacyl-AMP + PPi Aminoacyl-AMP + tRNA ↔ Aminoacyl-tRNA + AMP • • • • Each amino acid has „its own“ tRNA and aminoacyl-tRNA synthetase (ARS) Reactions are cytosolic Errors are corrected by specific correcting enzymes ARS have other enzyme activity (other signaling molecule?) Other ARS functions Proteosynthesis Iniciation Elongation Termination Post-translation processing of proteins • • • • Secondary structure and role of the chaperons Proteolytic modifications Glycosylation Other modifications (hydroxylation, phosphorylation, acetylation, methylation, carboxylation) Protein transfer after translation Contranslational translocation Contranslational translocation –transmembrane proteins Transmembrane proteins - examples • Type I – glycophorin, LDL receptor, influenza HA protein, insulin receptor, growth hormone receptor … • Type II – transferrin receptor, influenza HN protein, Golgi sialyltransferase, Golgi galactosyltranferase … • Type III – cytochrome P450 … • Type IV – G-protein, glucose receptors (GLUT 1 …), connexin, voltage gated Ca2+ channel … Secondary structure Secondary structure –HSP70 chaperon cycle Secondary structure – GroEL/GroES system Secondary structure – overview Protein disulfid isomerase (PDI) and peptidyl prolyl cis-izomerase PPI: Proteolytic modification - insulin N-Glycosylation Other modifications • O-glycosylation • Hydroxylation (hydroxyproline, hydroxylysine) • Methylation (mono- , di- and even trimethyllysine) • PHOSPHORYLATION • Carboxylation (γ-carboxyglutamate, vitamin K, fibrinogen) • Acetylation • …….. Protein degradation • • • • • Proteases Protein degradation systems Ubiquitin and proteasome Activation of proteases Protease inhibitors Proteases • Serine proteases (trypsin, chymotrypsin, elastase ….) • Aspartate proteases (pepsin, some proteases found in lysosomes, renin, HIV-protease …) • Metalloproteases (carboxypeptidases, various matrix metalloproteases …) • Cysteine proteases (papain, cathepsins, caspases, calpains …) Protein degradation systems • Vacuolar (lysosomes, endosomes, ER, …) • Ubiquitin pathway (proteasome) Ubiquitin pathway Activation of proteases • Most proteases are synthesized as larger pre-proteins. During activation, the pre-protein is cleaved to remove an inhibitory segment. • In some cases activation involves dissociation of an inhibitory protein • Activation may occur after a protease is delivered to a particular compartment within a cell or to the extracellular milieu. • Caspases involved in initiation of apoptosis are activated by interaction with large complexes of scaffolding and activating proteins called apoptosomes. Protease inhibitors • IAPs are proteins that block apoptosis by binding to and inhibiting caspases. The apoptosis-stimulating protein Smac antagonizes the effect of IAPs on caspases. • TIMPs are inhibitors of metalloproteases that are secreted by cells. A domain of the inhibitor protein interacts with the catalytic Zn2+. • Cystatins are inhibitors of lysosomal cathepsins. Some of these (also called stefins) are found in the cytosol and others in the extracellular space. Cystatins protect cells against cathepsins that may escape from lysosomes. • Serpins are widely distributed proteins that utilize a unique suicide mechanism to inhibit serine or cysteine proteases. A large conformational change in the serpin accompanies cleavage of its substrate loop. This leads to disordering of the protease active site, preventing completion of the reaction. The serpin remains covalently linked to the protease as an acylenzyme intermediate. • Non-specific: α2-macroglobulin Fate of the protein Literature • Marks´ Basic Medical Biochemistry, A Clinical Approach, third edition, 2009 (M. Lieberman, A.D. Marks) • B. Wilkinson, H.F. Gilbert / Biochimica et Biophysica Acta 1699 (2004) 35–44 • F. Ulrich Hartl, Andreas Bracher & Manajit HayerHartl, Molecular chaperones in protein folding and proteostasis, Nature 475 (2011)