* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download P0196 Poster Session I Basic science: pathogenesis of
Oncogenomics wikipedia , lookup
X-inactivation wikipedia , lookup
Non-coding DNA wikipedia , lookup
RNA interference wikipedia , lookup
Whole genome sequencing wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Human genome wikipedia , lookup
Molecular Inversion Probe wikipedia , lookup
History of genetic engineering wikipedia , lookup
Gene desert wikipedia , lookup
Copy-number variation wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
RNA silencing wikipedia , lookup
Public health genomics wikipedia , lookup
Non-coding RNA wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Genomic library wikipedia , lookup
Genome (book) wikipedia , lookup
Minimal genome wikipedia , lookup
Ridge (biology) wikipedia , lookup
Genome editing wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Microevolution wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Genomic imprinting wikipedia , lookup
Designer baby wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Gene expression programming wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genome evolution wikipedia , lookup
Metagenomics wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Pathogenomics wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
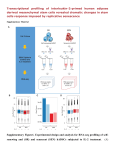
P0196 Poster Session I Basic science: pathogenesis of staphylococci COMPARISON OF TRANSCRIPTOMICS TECHNOLOGIES FOR ASSESSMENT OF STAPHYLOCOCCUS AUREUS GENE EXPRESSION D. Hernandez1, C. Le Priol2, D. Baud1, A. Fischer1, F. Reynier2, G. Gervasi2, S. Chatellier2, J. Veyrieras2, P. François1, J. Schrenzel1 1 Genomic Research Laboratory Division of Infectious Diseases, Geneva University Hospitals, Geneva, Switzerland 2 Technology Research Department Innovation and Systems Unit, BioMérieux, Marcy l'Etoile, France Objectives Today, high-throughput shotgun sequencing of transcriptomes (RNA-seq) in prokaryote seems to be an appealing alternative to well-established transcriptomics technologies such as microarray. While this later technology provides an analogical quantification of individual genes transcription (via the fluorescent intensity measuring the amount of hybridization between capture probes and their complementary cDNA fragments), RNA-seq methods make it possible to get a comprehensive digital quantification of transcribed regions (as done by counting the number of sequenced reads that map onto the corresponding genomic regions). Besides, contrary to existing digital technologies like the NanoString nCounter platform (and contrary to microarrays too), RNA-seq approaches do not require the prior design of probes and can then be used to get simultaneously the transcriptomic profile of prokaryote strains at both known and unknown transcribed regions. Nevertheless, analytical performances of RNA-seq approaches in prokaryotes have not been yet so far investigated. Here, we compared two RNA-seq solutions (Illumina MiSeq and Ion Torrent PGM) with Agilent microarrays and the NanoString nCounter system from Staphylococcus aureus total RNA samples. Methods We extracted four total RNA samples from the Staphylococcus aureus strain NCTC 8325. Samples were obtained at 3h and 5h of growth from a wild-type strain, as well as from a GdpS mutant. Each sample has been depleted from structural RNAs by using MicrobEnrich method (Ambion). The samples were then subjected to the different methods. RNA-seq data were mapped onto the reference genome sequence using BWA and converted to gene counts using Bedtools software applications. Statistical analysis was performed using the software R, with both the DESeq R package, as well as home-made scripts. Results Both Illumina and Ion-Torrent RNA-seq experiments displayed an average variation coefficient of about 25% between individual triplicates. However, at the gene level, the variation is strongly correlated with the individual coverage. Microarray and NanoString nCounter showed better reproducibility with Pearson correlation coefficients > 0.99. Conclusions RNA-seq, which is likely to become the standard approach in prokaryote transcriptomics, requires sufficient coverage for the results to be reliable. Since individual gene counts are not independent, highly expressed genes are detected at the expense of weakly covered genes for which reads counts may be insufficient for a reliable expression measurement. Both sequencing technologies are affected by sequence-related biases (such as %GC content), which may prevent comparing expression levels between individual genes. However, the sequence bias is strongly correlated for each sequencing technologies, which allows for differential expression measurements. The probebased NanoString nCounter system provides the most accurate expression measure and remarkable correlation between replicates. However, it only allows querying for a number of 800 targeted genes whose sequences have to be known.