* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download interrpo_nov16
Circular dichroism wikipedia , lookup
Rosetta@home wikipedia , lookup
Protein folding wikipedia , lookup
Protein design wikipedia , lookup
Bimolecular fluorescence complementation wikipedia , lookup
List of types of proteins wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein purification wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein domain wikipedia , lookup
Western blot wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein structure prediction wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Protein mass spectrometry wikipedia , lookup
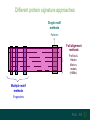
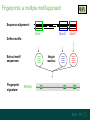
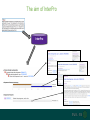
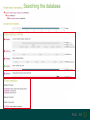
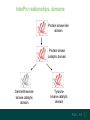
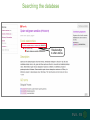
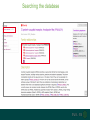
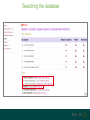
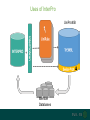
Understanding families, domains and function using InterPro Alex Mitchell [email protected] Overview • Why do we need tools to annotate protein function? • Ways in which we can transfer annotation: pairwise approaches signature based-approaches • What can we predict using these techniques? • Real world application of this knowledge to sequence data In an ideal world… • All proteins would be expressed and characterised in laboratories • Their sequences, structures and functions would be determined experimentally • This information would be stored, in a standardised human readable and computable form, in a freelyaccessible database The Swiss-Prot database Essentially, this was the idea behind the Swiss-Prot database: • Created in 1986 by Amos Bairoch • Contains protein sequence data which is manually annotated and reviewed • Information about protein function, structure, interactions, etc, extracted from the scientific literature by expert curators • Steadily grown to ~550k entries over 30 years Difficulties in experimentally characterising proteins • Can take many years in the lab to identify protein function • Lab experiments are expensive • Assays don’t exist for all functions • Difficulty in expressing some proteins Do something! Falling sequencing data costs Wetterstrand KA. DNA Sequencing Costs: Data from the NHGRI Large-Scale Genome Sequencing Available at: www.genome.gov/sequencingcosts. Growing sequence data volumes Growing sequence data volumes Making sense of sequence data Sequence data generation is no longer a major challenge in biology We have bucket loads of protein sequences (literally) • 10s of millions of sequences from over ½ a million species already in the databases • Can produce 100s of millions of predicted protein sequences from a single run on a sequencing machine Making sense of sequence data Inferring information from other proteins - BLAST • BLAST: Basic Local Alignment and Search Tool Pairwise comparison of proteins Widely used User friendly • Very good at recognising similarity between closely related sequences Using BLAST for annotation Using BLAST for annotation Using BLAST for annotation Using BLAST for annotation • Issues of scalability (comparison of 1 sequence against UniProt requires ~ 60 million pairwise comparisons) • Issues of sensitivity (detection of distant homologues) Using BLAST for annotation BLAST alignment of 2 proteins: • 60S acidic ribosomal protein P0 from 2 closely-related species 60S acidic ribosomal protein P0: multiple sequence alignment Protein signatures Alternatively, model the pattern of conserved amino acids at specific positions within a multiple sequence alignment • Patterns • Profiles • Profile HMMs Use these models (signatures) to infer relationships with the characterised sequences from which the alignment was constructed Approach used by a variety of databases: Pfam, TIGRFAMs, PANTHER, Prosite, etc ,,, Different protein signature approaches Single motif methods Patterns Full alignment methods Profiles & Hidden Markov models (HMMs) Multiple motif methods Fingerprints Patterns Many important sequence features, such as binding sites or the active sites of enzymes, consist of only a few amino acids that are essential for protein function Sequence alignment: Motif Extract pattern sequences: Build regular expression: Pattern signature: ALVKLISG AIVHESAT CHVRDLSC CPVESTIS [AC] – x -V- x(4) - {ED} PS00001 Fingerprints: a multiple motif approach Sequence alignment: Motif 1 Motif 2 Motif 3 Define motifs: Extract motif sequences: Fingerprint signature: xxxxxx xxxxxx xxxxxx xxxxxx PR00001 Weight matrices xxxxxx xxxxxx xxxxxx xxxxxx xxxxxx xxxxxx xxxxxx xxxxxx Fingerprints • Very good at modeling the often small differences between closely related proteins • Can distinguish individual subfamilies within protein families, allowing functional characterisation of sequences at a high level of specificity Fine-grained analyses Profiles & HMMs Whole protein Sequence alignment: Define coverage: Use entire alignment of domain or protein family Build model: Profile or HMM signature: Entire domain xxxxxxxxxxxxxxxxxxxxxxxxx xxxxxxxxxxxxxxxxxxxxxxxxx xxxxxxxxxxxxxxxxxxxxxxxxx xxxxxxxxxxxxxxxxxxxxxxxxx xxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxx xxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxx xxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxx xxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxx Profiles Start with a multiple sequence alignment Amino acids at each position in the alignment are scored according to the frequency with which they occur Scores are weighted according to evolutionary distance using a BLOSUM matrix Good at identifying homologues HMMs Start with a multiple sequence alignment Occupancy and amino acid frequency at each position in the alignment are encoded Insertions / deletions can be modelled Best path calculated Can model very divergent regions of alignment Very good at identifying evolutionarily distant homologues Homology search sensitivity Functional inference However, homology does not necessarily imply conserved function GHMP kinase superfamily includes: • Galactokinases (EC 2.7.1.6) • Homoserine kinases (EC 2.7.1.39) • Mevalonate kinases (EC 2.7.1.36) • Diphosphomevalonate decarboxylases (EC 4.1.1.33) Protein signatures • Different methodologies with different areas of application • A range of different sequence analysis databases that use these different methods exist • Combining these databases together would allow us to capitalise upon their individual strengths • This was the idea behind InterPro InterPro - integrated classification of protein families The aim of InterPro InterPro Features of InterPro • Manually annotated with literature referenced abstracts and Gene Ontology terms • Public releases every 2 months • Manually checked and updated against the Swiss-Prot database • Errors are identified and fixed GO annotation in InterPro: why stability does not indicate accuracy in a sea of changing annotations Sangrador-Vegas et al., Database (2016) doi: 10.1093/database/baw027 Searching the database Searching the database Searching the database Type Name Identifier Contributing signature Annotation InterPro entry types Family Proteins share a common evolutionary origin, as reflected in their related functions, sequences or structure Domain Distinct functional, structural or sequence units that may exist in a variety of biological contexts Repeats Short sequences typically repeated within a protein Sites PTM Active Site Binding Site Conserved Site Searching the database Side menu Searching the database Searching the database Searching the database Type Annotation Searching the database Searching the database Type Annotation Searching the database Searching the database Searching the database Relationships to other entries InterPro relationships: families InterPro relationships: domains Protein kinase-like domain Protein kinase catalytic domain Serine/threonine kinase catalytic domain Tyrosine kinase catalytic domain Searching the database Relationships to other entries Searching the database Searching the database Searching the database Viral rhodopsin-like GPCRs? Searching the database Searching the database What have we learned? Built up a lot of information very quickly, based on the sequence alone Membrane protein with 7 TM structure Retinal binding site in the C-terminus Member of the rhodopsin-like GPCRs • Broad class of membrane-bound receptors • Involved in signal transduction via binding to G-proteins • Mainly found in eukaroytes, but certain viruses have co-opted them • Structure solved for some family members (and shown to dimerise) What have we learned? Built up a lot of information very quickly, based on the sequence alone More specifically, member of the red/green sensitive opsin subfamily: • Found only in animals • Expressed in cone cells in the eye • Involved in phototransduction and colour vision • Absorption maximums at 560 & 530nm light wavelengths • Mutations/deficiencies can cause types of colour blindness Bulk sequence analysis This is useful for detailed analysis of individual proteins But for bulk analyses? • Genomes? • Proteomes? • Comparative analyses? GO term annotation GO terms Functional annotation: Gene Ontology • Grew out of the model organism community • Aims to unify the representation of gene and gene product attributes across species • Allows cross-species and/or cross-database comparison http://geneontology.org/ The Gene Ontology Less specific concepts • A way to capture biological knowledge in a written and computable form • A set of concepts and their relationships to each other arranged as a hierarchy More specific concepts www.ebi.ac.uk/QuickGO The Concepts in GO • • 1. Molecular Function An elemental activity or task or job protein kinase activity insulin receptor activity 2. Biological Process A commonly recognised series of events • 3. Cellular Component Where a gene product is located cell division • mitochondrion • mitochondrial matrix • mitochondrial inner membrane InterPro GO terms • GO terms are manually assigned to InterPro entries, based on scientific literature • Sequences searched against the database are annotated with GO terms, depending on the entry they match InterPro InterPro entry specificity determines the GO terms assigned GO:0007186 G-protein coupled receptor signaling GO:0016021 integral to membrane GO:0007186 G-protein coupled receptor signaling GO:0016021 integral to membrane GO:0007601 visual perception InterPro GO terms Using GO’s standardised & controlled vocabulary allows largescale consistent & human readable annotation of data sets, enabling comparison Uses of InterPro UniProtKB Member Databases Uses of InterPro • InterPro is the largest source of GO terms in UniProt • Provides ~ 120 million GO terms to >40 million distinct UniProtKB sequences GO term Evidence Source Uses of InterPro Protein discovery, functional characterisation and protein family analysis Uses of InterPro Analysis & comparison whole genomes, proteomes & transcriptomes Uses of InterPro Analysis of metagenomes from environmental samples Summary InterPro allows scientists to quickly amass information about individual protein sequences: • the domains and sites that they contain • the families to which they belong • the species in which these type of proteins are found, the pathways they are involved in, GO term annotation, etc The database also allows bulk analysis of genomes, proteomes and metagenomes, enabling: • comparative analyses • data discovery for translational research Acknowledgements The Protein Families team: Hsin-Yu Chang Sara El-Gebali Aurelien Luciani Matthew Fraser Jaina Mistry Gift Nuka Sebastien Pesseat Simon Potter Matloob Qureshi Neil Rawlings Lorna Richardson Gustavo Salazar-Orejuela Amaia Sangrador Siew-Yit Yong Group Head: Rob Finn Cluster Head: Alex Bateman Hands-on • Exploring InterPro to find predicted information about protein sequences • Protein functional inference using InterPro and BLAST Mini group research project, using InterPro and other databases