* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Print
Survey
Document related concepts
Structural alignment wikipedia , lookup
Western blot wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Homology modeling wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein domain wikipedia , lookup
Protein folding wikipedia , lookup
Circular dichroism wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein mass spectrometry wikipedia , lookup
Transcript
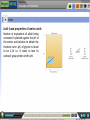
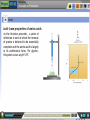
Proteomics Preamble to Proteomics Preamble to Proteomics The term “proteome” describes the protein complement expressed by a genome. The study of the full set of proteins encoded by a genome is known as proteomics. This large scale study of protein structure and function, first requires a thorough understanding of protein composition and their various structural levels. Learning Objective After interacting with this Learning Object, the learner will be able to, Recall Amino acids structures & classification. Describe acid-base properties of Amino-acids. Define Peptide bond formation, and, Recall protein structural levels. Proteomics Preamble to Proteomics Amino acid structures & classification Amino acids are the building blocks or monomers that make up proteins. They consist of a central alpha carbon atom bonded covalently to an amino group, a carboxyl group, a hydrogen atom and a variable side chain, also called the R group. While most amino acids have a primary amino group, proline consists of a secondary amine group and is therefore an imino acid. Proteomics Preamble to Proteomics Amino acid structures & classification All amino acids except glycine have a non-superimposable mirror image due to the spatial arrangement of four different groups about the chiral carbon atom. Rotation of either isomer about its central axis will never give rise to the other isomeric structure. Proteomics Preamble to Proteomics Amino acid structures & classification Amino acids are classified based on the properties of their side chains or R groups which vary in size, structure and charge. The polarity of the side chains is one of the main basis for classification. Proteomics Preamble to Proteomics Acid- base properties of amino acids All amino acids exhibit a characteristic titration curve with distinct pK values. Amino acid taken in an acidic medium is titrated against 0.1N NaOH in a burette. The cationic form of the amino acid is gradually converted into its neutral or zwitterionic form by loss of a proton from its COOH group. Proteomics Preamble to Proteomics Acid- base properties of amino acids Number of equivalents of alkali being consumed is plotted against the pH of the amino acid solution to obtain the titration curve. pK1 of glycine is found to be 2.34 i.e. it starts to lose its carboxyl group proton at this pH. Proteomics Preamble to Proteomics Acid- base properties of amino acids As the titration proceeds, a point of inflection is seen at which the removal of proton is believed to be essentially complete and the amino acid is largely in its zwitterionic form. For glycine, this point occurs at pH 5.97. Proteomics Preamble to Proteomics Acid- base properties of amino acids Removal of the proton from the amino group constitutes the second stage of the titration process. The zwitterionic form is gradually converted into the anionic form until the pH is sufficiently alkaline to contain amino acid only in the alkaline form. Proteomics Preamble to Proteomics Acid- base properties of amino acids The pK2 of an amino acid is obtained in the second stage of the titration. pK2 of glycine is found to be 9.6. Some amino acids having positively or negatively charged side chains will have pK1, pK2 and pKR, which corresponds to ionization of the side chain. These amino acids have good buffering capacity around 1 pH unit on either side of their pK values. Proteomics Preamble to Proteomics Peptide bond formation Amino acids are the building blocks or monomers that make up proteins. Proteomics Preamble to Proteomics Peptide bond formation Amino acids are oriented in a head-totail fashion and linked together such that the carboxyl group of one amino acid combines with the amino group of another. Two amino acids joined together by means of such a condensation reaction with the loss of a water molecule forms a dipeptide. Many such amino acids linked together form a polypeptide. Proteomics Preamble to Proteomics Peptide bond formation The peptide bond is rigid due to its partial double bond character. However, the bonds between the αcarbon and amino and carboxyl groups are pure single bonds that are free to rotate. Proteomics Preamble to Proteomics Protein structural levels Amino acids are joined together in a head-to-tail arrangement by means of peptide bonds with the release of water molecules. This linear sequence of amino acids constitutes the primary structure. Proteomics Preamble to Proteomics Protein structural levels The folding of the primary structure into the secondary is governed by the permissible rotations about the and ψ angles. Not all values of these angles lead to sterically favorable conformations. The Ramachandran’s plot defines the regions of favorability. Proteomics Preamble to Proteomics Protein structural levels Amino acids along the polypeptide backbone interact through hydrogen bonds leading to secondary structures. The -helix has intra-chain hydrogen bonds between the ‘H’ of NH and ‘O’ of CO in every 4th residue. In sheets, the backbone is made to zigzag such that chains are arranged side by side for hydrogen bonding. Proteomics Preamble to Proteomics Protein structural levels Amino acids located far apart on the polypeptide chain interact with each other by means of hydrogen bonds, electrostatic interactions, disulphide bridges etc., allowing the protein to fold three dimensionally in space, giving rise to the tertiary structure. Folding takes place such that the hydrophobic residues are buried inside the structure while the polar residues remain in contact with the surroundings. Proteomics Preamble to Proteomics Protein structural levels Different subunits or polypeptide chains interact with one another and are held together by means of ionic, electrostatic, van der Waals etc interactions. Such multisubunit proteins are said to have a quaternary structure. Proteomics Preamble to Proteomics Structure & Classification 1. Amino acid: The basic monomeric unit of polypeptides and proteins. There are twenty standard amino acids with different structures and properties that can be combined in multiple ways to make up the wide range of proteins known to us. Each amino acid is also specified by a three-letter and single letter code. 2. α-carbon atom: The central carbon atom of an amino acid which is covalently bonded to an amino group (NH2), a carboxyl group (COOH), a hydrogen atom (H) and a variable R group. The groups are tetrahedrally arranged around the carbon atom. This carbon atom is an asymmetric or chiral centre that gives rise to the phenomenon of optical isomerism thereby conferring a non-super imposable mirror image on each of the amino acids except glycine. 3. Side chain: The side chain or R group is distinct for each amino acid, giving them their unique properties. It is on the basis of this side chain that the amino acids are classified into various groups. 4. Amino group: This consists of an NH2 group covalently bonded to the central carbon atom. Depending upon the pH of the surrounding medium, it either exists as NH2 or NH3+ . Except for proline, which has a secondary amino group, all amino acids have only primary amino groups. 5. Carboxyl group: A COOH group covalently bound to the central alpha carbon atom, which exists as either COOH or COO- depending on the pH of the surrounding medium. Proteomics Preamble to Proteomics Acid-base properties 1. Cationic form: All amino acids exist in the completely protonated form in acidic medium, known as the cationic form. Both amino and carboxyl groups are protonated here. 5. Amino acid in acidic medium: To obtain the titration curve of an amino acid, it is first taken in a highly acidic medium such that it exists entirely in the cationic form. 2. Zwitterion: The state in which the amino acid has no net charge is known as the zwitterion. It is neutral due to the presence of equal number of NH3+ and COO- groups. 6. Burette: A graduated, long glass tube fitted with a stopcock at the end to control the flow of liquid. This contains the solution against which titration is to be performed. In this case, the amino acid is titrated against 0.1N sodium hydroxide (NaOH). 3. Anionic form: In a highly alkaline medium, all amino acids exist in their anionic form due to the presence of COO- group. 4. Titration flask: A conical flask in which the solution to be titrated is taken along with a suitable pH indicator. 7. Stand: This has a clamp which can hold the burette steady while the experiment is being performed. Proteomics Preamble to Proteomics Acid-base properties 8. Titration curve: The number of equivalents of alkali being consumed during the titration process is plotted against pH of the solution in the flask to yield a unique titration curve for each amino acid. The titration curve depicted corresponds to that of glycine. 9. pK: Negative log of the pH at which the catonic and neutral forms inter-convert (pK1) and neutral and anionic forms inter-convert (pK2). Proteomics Preamble to Proteomics Peptide bond 1. Peptide bond: The bond formed during the process of linking together two amino acids with the carboxyl group of one amino acid being linked to the amino group of another with the concurrent loss of a water molecule. These bonds are planar in geometry and exhibit partial double bond character. 2. Dipeptide: Two amino acids bonded through a peptide bond. Many such amino acids linked together constitute a polypeptide. 3.ψ (psi) and φ (phi): Angle of rotation about the bond between the α-carbon atom and carboxyl and amino groups respectively. These angles determine which protein conformations will be favourable. Proteomics Preamble to Proteomics Structural levels 1. Primary structure: The sequence of amino acids joined together by peptide bonds to form a linear polymer constitutes the primary structure of the protein. Linear polypeptide chains are often cross-linked, most commonly by two cysteine residues linked together to form a cystine unit. 2. Secondary structure: The folding of a polypeptide backbone by means of internal hydrogen bonds between nearby amino acid residues giving rise to a regular arrangement defines the secondary structure of the protein. α-helices and β-sheets are the most commonly observed secondary structures of proteins due to their highly favourably ψ and φ angles as described by the Ramachandran’s plot. The amino acid proline tends to disrupt the helix and is often found at a bend in the structure known as reverse turns or β bends. 3. Tertiary structure: Interactions (hydrophobic, electrostatic, hydrogen bonds etc.) between amino acid side chains located far apart in the polypeptide sequence cause the protein to fold resulting in a three-dimensional arrangement of atoms known as the tertiary structure. The folding takes place in such a way that the hydrophobic residues get buried to form the core while the hydrophilic amino acids remain on the surface in contact with the polar surroundings. 4. Quaternary structure: Many proteins have more than one polypeptide chain, also called a subunit, that are assembled together by various interactions like electrostatic, van der Waals, disulphide bonds etc. giving rise to the quaternary structure. Proteomics Preamble to Proteomics Structural levels 5. Bonding interactions: Several types of bonds are responsible for stabilizing protein structures. Some of them are: i) Hydrogen bonds: These are formed between an electronegative atom (like O or N) and a highly electropositive atom (like H). They can be formed within a polypeptide chain (intrachain), as in the case of secondary structures, or between different polypeptide chains (interchain). ii) Electrostatic interactions: Attractive forces existing between oppositely charged groups/atoms, which can stabilize the protein structure. iii) Hydrophobic interactions: These are largely non-specific interactions between nonpolar amino acid side chains, which act to bury these hydrophobic residues away from a polar environment. iv) Van der Waals forces: These are attractive or repulsive forces caused due to fluctuating polarization and therefore temporary dipole formation between nearby particles. v) Disulphide bridges: Specific interaction and oxidation of thiol groups of cysteine residues in different regions of the polypeptide chain(s) leads to formation of disulphide (S-S) bonds. Proteomics Preamble to Proteomics Books