* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Chapter 14 – From Gene to Phenoytpe
Epigenetics of diabetes Type 2 wikipedia , lookup
Genome evolution wikipedia , lookup
Gene desert wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
Frameshift mutation wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Gene therapy wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Genome (book) wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Protein moonlighting wikipedia , lookup
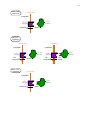
Site-specific recombinase technology wikipedia , lookup
Gene expression programming wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Gene expression profiling wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Designer baby wikipedia , lookup
Gene nomenclature wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Point mutation wikipedia , lookup
1 Chapter 14 – From Gene to Phenoytpe Questions to be addressed: • In typical crosses, what ratios are expected for genes with alleles that do not show complete dominance/recessive relationships? • What ratios are expected from crosses if more than one gene controls a particular characteristic? • What can these ratios tell us about the function of the genes? Terminology (see also Glossary pages 607-634 and web site http://helios.bto.ed.ac.uk/bto/glossary/) pleiotrophic mutation: a mutation that affects several different phenotypic characteristics codominance: alleles which, when combined in the heterozygote show aspects of both homozygotes incomplete dominance: alleles which, when combined in the heterozygote show a phenotype intermediate between the homozygotes epistasis: the phenotype of a mutant allele at one gene overrides the phenotype of a mutant allele at another gene, such that the double mutant has the same phenotype as the first mutant suppression: a mutation in one gene can cancel the effect of a mutation in a second gene, resulting in a wild type phenotype penetrance: the proportion of individuals with a specific genotype who show that genotype phenotypically expressivity: the degree to which a particular genotype is expressed in the phenotype Mendel’s findings: • one gene, two alleles • each gene controls a single different character 2 • one allele is dominant to the other ratios = = 3 : 1 = 1 : 2 : 1 (monohybrid) heterozygotes 9 : 3 : 3 : 1 (dihybrid) test cross of 1 : 1 (monohybrid) heterozygotes 1 : 1 : 1 : 1 (dihybrid) Complexity arises: • a gene may have multiple alleles: each gene consists of many nucleotides, which when mutated result in different alleles • • different alleles may have different phenotypes a gene may be pleiotrophic: a single gene acts to control more than one phenotypic characteristic • e.g. gene controlling pollen grain germination also controls root hair growth • suggests a common underlying physiological or developmental mechanism • more than one gene may control a single characterisic • since many biological pathways consist of a series of steps, each controlled by a different protein (coded by a gene) , a single characteristic may be the result of multiple gene activity 1. Each gene can have multiple alleles • How do you know if 2 mutant lines with the same phenotype are the result of: a) mutations in 2 genes in the same pathway b) 2 alleles of the same gene • example: consider the trp biosynthetic pathway 3 trpA+ X trpB+ Y trpC+ tryptophan experiment: two new auxotrophic mutants for tryptophan are found in the plant Arabidopsis thaliana hypothesis 1: the two mutants are alleles of same gene, A (e.g. mutant 1 = a1, mutant 2 = a2) hypothesis 2: the two mutants are alleles of different genes A and B (e.g. mutant 1 = a1, mutant 2 = b1) experiment: cross two mutants together and the results will differ depending on which hypothesis is correct expected results for hypothesis 1 conclusion: F1 progeny is mutant, therefore mutations do not complement one another, and the mutations are alleles of the same gene expected results for hypothesis 2 conclusion: F1 progeny is wild type, therefore two mutants complement one another, and must be mutations in different genes RULES FOR A COMPLEMENTATION TEST • can only be done with recessive mutations 4 • If the mutations are in different genes, the two mutations will complement one another (progeny will be wild type) • If the mutations are alleles of the same gene, the two mutations will not complement one another (progeny will be mutant) VARIATIONS ON THE COMPLEMENTATION TEST – THE HETEROKARYON (figure 6-2) • in organisms with a haploid life cycle, cannot do a complementation test • instead, fuse two cells • the resulting cells have 2 nuclei = heterokaryon in a common cytoplasm • gene products from both nuclei act within the common cytoplasm Dominance relationships amongst alleles Most frequently •dominance = presence of enzyme function A -----> functional enzyme • recessiveness = absence of enzyme function a -----> no functional enzyme in Aa, presence of A (functional enzyme) will override presence of a (nonfunctional enzyme), therefore A will be dominant to a Aa = A phenotype Example 1 Phenotype depends on a threshold amount of protein. eyeless + (ey+ )- required to form eyes in Drosophila -each ey+ allele makes 50 units protein/cell therefore, ey+ ey+ - makes 100 units of protein/cell 5 Case I -a cell needs 80 units of Ey functional protein to form an eye -mutant allele ey-1 makes 0 units (a null allele, no protein is made) -thus a homozygous mutant (ey-1ey-1) fly has no eye What is the phenotype of an ey+ ey-1 fly? Case 2 a cell requires 80 units of Ey protein to form an eye allele ey-2 makes 35 units of protein (a partial loss of function) -cells in a homozygous mutant (ey-2 ey-2) fly make 70 units, therefore this fly has no eye Example 2 The phenotype varies with the concentration of protein. allele ELF+ - necessary to delay flowering (ELF = early flowering) - makes 50 units protein/cell ELF+ ELF+ - produces 100 units protein cell (makes flowers in 24 days) allele elf - makes 0 units protein/cell elf elf - produces 0 units protein/cell ELF+ elf - produces 50 units/cell (flowers in 0 days) (flowers in 12 days) -the heterozygote is intermediate between two homozygous individuals = INCOMPLETE DOMINANCE 6 Question: if heterozygotes are crossed, what ratio is expected in the progeny? Conclusion: If the heterozygote has a phenotype distinct from either homozygote, the phenotypic ratio will be the same as the genotypic ratio. Dihybrid crosses involving altered dominance/recessive relationship Trait 1 Trait 2 ELF+ ELF+ = flowers in 24 days ELF+ elf = flowers in 12 days elf elf = flowers in 0 days RR = red Rr = red rr = white CROSS = ELF+ elf Rr X progeny? (3:1 x 3:1 = 9:3:3:1?) ELF+ elf Rr 7 3/4 R_ 3/16 ELF+ELF+R_ 1/4 rr 1/16 ELF+ELF+rr 3/4 R_ 6/16 ELF+elf R_ 1/4 rr 2/16 ELF+elf rr 3/4 R_ 3/16 elf elf R_ 1/4 rr 1/16 elf elf rr 1/4 ELF+ ELF+ 1/2 ELF+ elf 1/4 elf elf ratio = 3 : 1: 6 : 2 : 3 : 1 (= 1:2:1 x 3:1) Example 3 What if alleles produce proteins having different and independent functions? Blood groups • blood is categorized into different types based on how blood reacts to a set of antibodies • each antibody interacts with a particular antigen (protein) present on the cell surface • the presence of such an interaction is recognized by blood clumping • one antigen (protein) is produced by gene I • -3 alleles (IA, IB, i) which produce different antigens recognized by different antibodies IA reacts with A antibody I B reacts with B antibody i - reacts with no antibody (null allele) genotype antigen blood group IA IA IB IB A B A B ii none O 8 What if we consider heterozygous combination? genotype antigen blood group IA I B IB i A and B B AB B IA i A A CODOMINANT - heterozygote has phenotypic characteristics of both homozygotes therefore, IA and IB are codominant to one another IA is dominant to i I B is dominant to I Question? A baby has blood type O. Two couples claim to be the baby’s parents: • couple 1 has blood groups A and B • couple 2 has blood groups AB and O Which are the parents, couple 1 or couple 2? conclusion: 9 Ambiguity in terminology e.g. sickle cell anemia, a tropical disease caused by a mutation in the gene coding for hemoglobin HbA HbA -wild type, blood cells round, NO anemia HbS HbS - blood cells sickle shaped, ANEMIC HbA HbS - blood cells normal, except under low O2, NO anemia extract hemoglobin and electrophorese Hemoglobin types present S and A S A Genotype Hb S Hb A Hb S Hb S Hb A Hb A Phenotype Sickle-cell / Normal cell Sickle-cell Normal shape Origin Positions to which hemoglobins have migrated (see Figure 14-8) If we consider different phenotypic characteristics, the dominance/recessive descriptions vary: phenotypic characteristic = disease phenotypic characteristic = migration phenotypic characteristic = cell shape under low O2 10 Lethal Alleles e.g. manx cat X manx cat (manx = talless due to abnormal spinal development, Figure 14-11) results: progeny = 2 manx cat and 1 tailed cat Conclusion: manx cats are not true breeding, moreover, 2:1 does not look like a standard Mendelian ratio Simplest hypothesis: this is a Mendelian ratio, but one class is combined with another or missing (a modified Mendelian ratio) - what standard ratios might we consider for such a modification? hypothesis: wild type (tailed) = manx (tailless) = experiment: cross 2 manx cats and look for lethality during embryogenesis results: 1/4 of embryos have arrested development (lethal) conclusion: M+ is necessary for spinal cord development M M -> die early in embryonic development due to spinal cord malformation M+ M - have sufficient M+ protein to make most of the spinal cord, but the tail is absent Part 4: Genes controlling same characteristic 11 example 1: Recessive epistasis -the dark red colour of wild type Drosophila eyes results from a biochemical pathway with two steps, each controlled by a different gene (v+ and s+) v+ vermillion s+ scarlet dark red recessive allele v (vv flies = vermilion) recessive allele s (ss flies = scarlet) CROSS: vermillion fly X scarlet fly PARENTAL: vvs+s+ X v+v+ss v+vs+s (wildtype) 9 v+_ s+_ 3 v+ _ ss 3 vv s+_ 1 vvss F1: F2: (wild type) (scarlet) (vermillion) (?) recall the pathway v+ vermillion s+ scarlet dark red What colour will flies nonfunctional at both v+ and s+ be? ratio = • we say that phenotype v (vermilion) is epistatic to phenotype s (scarlet) (epistatic = stands over) 12 NOTE: if one gene is epistatic to another, it usually means that the epistatic gene acts first (is upstream) in the biochemical (or developmental) pathway, and that the two genes act independently (on different steps within the same pathway example 2: Complementary gene action • mutations in either gene gives the same phenotype trpA+ X trpB+ Y trpC+ tryptophan trpC+ mutant allele c - auxotrophic for trp trpB+ mutant allele b - auxotrophic for trp ratio = Example 3: Suppression • a mutation in one gene compensates for a mutation in a second gene • good evidence that two gene products interact A_B_ = aaB_ = aabb = A_bb = b suppresses (compensates for) a 13 Example: protein A + protein B functional protein (dimer) -a protein containing 2 different subunits = heterodimer -a protein containing 2 identical subunits = homodimer clavata mutants = clv1 and clv3 mutation in CLV1 (clv1) causes the division of cells in apical meristem to become uncontrolled ----> enlarged and club-shaped apex mutation in CLV3 (clv3) also causes the division of cells in apical meristem to become uncontrolled -----> enlarged and club-shaped apex 14 WILD TYPE response membrane cytoplasm CLV3 protein CLV1 protein response signal MUTANT responses membrane membrane cytoplasm cytoplasm CLV3 protein clv1 protein NO response NO response signal WILD TYPE response membrane cytoplasm clv3 protein clv1 protein response signal clv3 protein CLV1 protein NO signal 15 phenotypic ratio = What if there was an allele of CLV3 (clv3*) which could bind with both wild type CLV1 and mutant clv1? CLV1CLV1CLV3CLV3 = CLV1CLV1clv3*clv3* = clv1clv1 CLV3CLV3 = clv1clv1clv3*clv3* = phenotypic ratio = Example 4: Genetic Redundancy (duplicate genes) (two genes doing the same thing – loss of one gene can be compensated for by presence of other gene) chemotaxis in Caenorhabditis elegans (a nematode) WAN1 and WAN2 - both expressed in sensory neurons wan1 - mutation causing lack of WAN1 protein wan2 - mutation causing lack of WAN2 protein WAN1WAN1 WAN2WAN2 -moves to chemical (wild type response) wan1wan1 WAN2WAN2 - moves to chemical WAN1WAN1 wan2wan2 - moves to chemical wan1wan1wan2wan2 - wanders 16 -wild type copy of one WAN gene can compensate for loss of the other WAN gene -only see a phenotype if both functions are lost RATIO = MODIFIED DIHYBRID RATIOS RESULTING FROM GENE INTERACTIONS Type of gene interaction 9 A_B_ 3 A_bb 1 aabb Phenotypic ratio 3 1 9:3:3:1 None (four discrete phenotypes) 9 Complementary gene action 9 Supression 9 3 4 13:3 9 3 4 9:3:4 Recessive epistasis of aa action on B and b alleles Dominant expistasis of 3 3 aaB_ 9:7 7 12 3 1 12:3:1 1 15:1 A acting on B and b alleles Duplicate genes 15 Haploid Organisms 1) interaction between alleles (e.g. dominance/recessive relationships) -can only define if phenotype is expressed in the diploid stage 2) interaction between genes 17 (epistasis, suppression, redundancy) example: genes controlling spore (haploid) pigmentation in Neurospora (fungus) al = albino al+ = orange ylo = yellow ylo+ = orange conclusion: al is epistatic to ylo, suggesting the following biosynthetic pathway Penetrance and Expressivity (Figure 14-22) PENETRANCE - the percentage of individuals with a given genotype who exhibit the phenotype associate with that genotype B_ = normal vision bb = blind but, of 100 people of genotype bb, only 20 are blind therefore, allele b has a penetrance of 20% 18 EXPRESSIVITY - the extent to which a given phenotype is expressed in an individual A_ = normal pigmentation aa = albino -one individual has genotype aa, but has 30% of normal skin pigmentation