* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Detection of unpaired DNA at meiosis results in RNA‐mediated
Genomic library wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Pathogenomics wikipedia , lookup
Gene desert wikipedia , lookup
Gene therapy wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Genomic imprinting wikipedia , lookup
Genetic engineering wikipedia , lookup
Gene expression programming wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Epitranscriptome wikipedia , lookup
Transposable element wikipedia , lookup
Human genome wikipedia , lookup
History of RNA biology wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Oncogenomics wikipedia , lookup
X-inactivation wikipedia , lookup
Minimal genome wikipedia , lookup
Genome (book) wikipedia , lookup
Non-coding DNA wikipedia , lookup
Gene expression profiling wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
History of genetic engineering wikipedia , lookup
Non-coding RNA wikipedia , lookup
Nutriepigenomics wikipedia , lookup
RNA interference wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Primary transcript wikipedia , lookup
Genome editing wikipedia , lookup
Designer baby wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Genome evolution wikipedia , lookup
Point mutation wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Helitron (biology) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
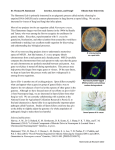
What the papers say Detection of unpaired DNA at meiosis results in RNA-mediated silencing Michael J. Hynes* and Richard B. Todd Summary During meiosis, homologous chromosomes must pair in order to permit recombination and correct chromosome segregation to occur. Two recent papers(1,2) show that meiotic pairing is also important for correct gene expression during meiosis. They describe data for the filamentous fungus Neurospora crassa that show that a lack of pairing generated by ectopic integration of genes can result in silencing of genes expressed during meiosis. This can result in aberrant meioses whose defects are specific to the function of the unpaired gene. Furthermore, mutations affecting the silencing mechanism have been selected in a gene encoding a putative RNAdependent RNA polymerase. This finding indicates the involvement of a meiotic specific post-transcriptional gene silencing mechanism (PTGS) similar to that observed in vegetative cells in N. crassa and other organisms. Finally, this gene product is essential for normal meiosis, suggesting that RNA-dependent processes are fundamental to the sexual cycle. BioEssays 25:99–103, 2003. ß 2003 Wiley Periodicals, Inc. Introduction The involvement of small noncoding RNA molecules in a variety of biological processes has recently excited considerable attention. Amongst these are microRNAs (miRNAs), about 22–25 nucleotides long, generated from longer precursor molecules by the action of the Dicer class of enzymes.(3–5) These are generally involved in regulating mRNA stability and translation and include molecules (small Department of Genetics, University of Melbourne, Australia. Funding agency: Sequencing of A. fumigatus was funded by the National Institute of Allergy and Infectious Disease U01 AI 48830 to D. Denning and W. Nierman. *Correspondence to: Prof. Michael J. Hynes, Department of Genetics, University of Melbourne, Parkville 3010, Australia. E-mail: [email protected] DOI 10.1002/bies.10241 Published online in Wiley InterScience (www.interscience.wiley.com). Abbreviations: PTGS, post-transcriptional gene silencing; miRNAs, microRNAs; stRNAs, small temporal RNAs; siRNAs, small intermediate RNAs; RdRP, RNA-dependent RNA polymerase; RIP, Repeat Induced Point mutation; MSUD, Meiotic Silencing by Unpaired DNA. BioEssays 25:99– 103, ß 2003 Wiley Periodicals, Inc. temporal RNAs—stRNAs) that are involved in normal control of development as exemplified by the lin4 and let7 genes of Caenorhabditis elegans.(6–9) The other general class are the small intermediate RNAs (siRNAs), derived from abnormal RNA, that lead to targetted mRNA destruction (post-transcriptional gene silencing- PTGS) in response to sensing foreign nucleic acids. A common theme of such silencing events appears to be the synthesis of double stranded RNA by RNAdependent RNA polymerase (RdRP).(10,11) The fungus Neurospora crassa apparently has an intense dislike for aberrant or duplicated nucleic acids and has two well-characterised mechanisms to regulate expression of foreign or duplicated sequences.(10,12) Studies of these mechanisms have made significant contributions to the field of gene silencing. Two recent papers describe a novel mechanism that senses unpaired DNA at meiosis resulting in silencing of genes expressed during meiosis.(1,2) If the gene is required for a meiotic event then meiosis will halt at a characteristic stage. Silencing in Neurospora crassa N. crassa provides an excellent system for studying the genetics, cytology and cell biology of meiosis.(13) Mating occurs between strains carrying alternate mating type alleles mat A and mat a, which are dissimilar in sequence (idiomorphs),(14) and results in the formation of premeiotic ascogenous hyphae in which nuclei fuse to form a diploid nucleus (karyogamy). This is followed by immediate meiotic divisions and one mitosis to yield eight nuclei, which are then enclosed into ascospores linearily arranged in an ascus (Fig. 1). The results of individual meioses can be observed by ascospore pigmentation or morphology, and the fertility of crosses assessed by ascospore yield and viability. In addition, N. crassa is readily transformed by DNA carrying selectable markers yielding transformants with DNA integrated either by homology or ectopic integration at nonhomologous sites. Two silencing mechanisms have previously been found in this organism. One (Repeat-Induced Point mutation—RIP) involves sensing duplicated sequences in the genome and results in multiple C to T mutations in these sequences, in a process specifically occurring in premeiotic cells.(12) Silencing, therefore, is permanent and is observed only following crossing. The second (quelling) detects multiple sequences in the genome in vegetative hyphae (e.g. those generated by BioEssays 25.2 99 What the papers say Figure 1. Silencing mechanisms occur in Neurospora crassa at various stages of the life cycle. Neurospora crassa grows as haploid multinucleate vegetative hyphae (A) Silencing of sequences present in multiple copies occurs in vegetative tissue by Quelling. Mating occurs only between two individuals of opposing mating type (mat A or mat a). Each individual can act as the female and can be fertilized by male elements of the other strain. The haploid nuclei derived from each strain proliferate in premeiotic dikaryotic ascogenous hyphae (B) by conjugated nuclear divisions. It is within this dikaryotic tissue that duplicated sequences are silenced by RIP (Repeat Induced Point-mutation). The hyphal tip forms a crozier, the terminal pair of nuclei undergo one synchronous mitotic division and the products are partitioned into different cells such that the penultimate cell contains one nucleus from each parent. These nuclei fuse (karyogamy) to form the diploid zygote (C), which immediately undergoes meiosis. The four meiotic progeny divide once mitotically and the resulting eight nuclei are packaged into ascospores arranged linearly within an ascus (D). During meiosis silencing of unpaired sequences occurs by MSUD (meiotic silencing by unpaired DNA). MSUD is observable in individual asci, with the specific phenotype dependent on the meiotic or ascopore development function of the silenced gene. Ascus development is accompanied by the development of a fruiting body (perithecium) (E), which contains all of the asci arising from a particular mating. transforming DNA) and silences expression of these by targetting the encoded mRNA for degradation.(10,15) The isolation of quelling deficient mutants clearly indicates a mechanism with similarities to PTGS mechanisms in other systems involving 25 nucleotide long siRNA production from aberrant transcripts and their amplification via RdRP.(16,17) Meiotic silencing The new mechanism was discovered via studies of a gene (asm-1) required for mature melanized ascospore production.(18) Deletions of the gene are dominant resulting in all ascospores in almost all asci being unmelanized and inviable. Ectopic copies of a wild type gene cannot repair the defect unless the cross is homozygous for the ectopic copy.(18) A requirement for pairing suggested a mechanism formally 100 BioEssays 25.2 related to the classical transvection phenomena of Drosophila. However the finding that crosses with two paired and one unpaired asm-1 copies result in virtually complete loss of ascospore maturation indicates that it is the unpaired copy that triggers silencing both of itself and the paired copies.(1) Consequently this phenomenon of ascus dominance has been termed Meiotic Silencing by Unpaired DNA (MSUD). A number of other ascus-dominant mutations which affect ascospore shape (Round spore), incorporation of nuclei into ascospores (Banana) and linear ascospore arrangement (Peak) also apparently result from MSUD.(1) In the latter case most peak alleles are recessive but the one dominant allele results from a translocation break point in the gene and hence disrupts correct pairing. Furthermore studies with ectopic and homologously integrated cloned genes such as the recA/ What the papers say RAD51 orthologue and those encoding products predicted to be required during meiotic divisions such as actin, b-tubulin and histones clearly show MSUD resulting in characteristic blocks in various stages of meiosis or spore maturation.(1) In contrast, for metabolic genes essential during vegetative growth but not during meiosis MSUD cannot be detected. It can be noted that investigation of the specific MSUDgenerated block observed for meiotic genes of unknown function provides a valuable tool for genetic dissection of meiosis. The mat A and mat a alleles are not expected to pair as they are dissimilar in sequence, and yet their expression is required during meiosis. Therefore they are likely to be immune to MSUD probably due to a special feature of their flanking sequences. Consistent with this interpretation, matings between a mat a strain and one with mat A integrated ectopically results in few ascospores.(1) Mutations affecting silencing Mutations affecting MSUD have been isolated(2) by selecting for survivors of a cross between an Asm-1 deletion mutant and a mutagenized strain with a wild type copy and an ectopic copy of a frameshifted asm-1 mutant gene. These were individually tested for suppression of MSUD by crossing to Round spore and looking for normal ascospore shape in the progeny. An initial mutant, Sad-1UV (suppressor of ascus dominance), was found to result in about one third normal ascospores in a heterozygous Round spore cross, with all eight spores in an ascus being normal. Sad-1UV does not suppress the Round spore phenotype itself because homozygous Round spore crosses yield 100% abnormal spores. The general suppression of MSUD by Sad-1UV has been demonstrated by suppression of the ascus dominance of Peak and Banana as well as restoring fertility to crosses with unpaired ectopicallyintegrated actin, histone, b-tubulin, mating type and RecA/ RAD51 genes.(1) Strains with duplications of chromosomal segments are barren, producing normal perithecia but no asci (or few), in crosses to wild type strains. Heterozygous crosses of Sad-1UV to such duplications showed increased fertility were fertile(1) consistent with the hypothesis that MSUD resulting from unpaired meiotic genes in duplicated segments is at least partly responsible for the dominant barren phenotype. Furthermore, crosses between N. crassa and some closely related species are infertile to varying degrees and Sad-1UV was found to increase fertility in these interspecies crosses.(1) This suggests that interspecies infertility at least in part results from numerous chromosomal rearrangements leading to lack of pairing of gene sequences that include genes essential for meiosis. If MSUD is present in many organisms, then this might constitute an important isolation mechanism for speciation. As selected, the Sad-1UV allele is partially dominant in heterozygous crosses. The Sad-1UV allele is deleted for the 30 half of the gene and contains a few RIP mutations in the 50 half.(1) Homozygous Sad-1UV crosses are barren with all asci arrested at meiotic prophase. This phenotype allowed mapping to linkage group I and cloning of sad-1þ by complementation.(2) It also has provided a more efficient method for the isolation of further Sad-1 mutations. A complete sad-1 deletion yielded almost 100% dominance while a series of sad-1 alleles generated by the RIP procedure gave varying levels of dominance correlating with both the number of induced base-pair changes and amounts of DNA methylation throughout the sequence even though all were mutated in the start codon.(1,2) DNA methylation is unlikely to be involved in triggering MSUD since dim-2 or rid mutations, which result in a lack of cytosine DNA-methyltransferases, do not suppress MSUD.(2) The varying levels of dominance of sad-1 alleles in heterozygous crosses can therefore be explained by autogenous MSUD leading to a low level of expression of sad-1þ generated by lack of pairing at meiosis. This illustrates a use for MSUD in analysing the stringency of requirements for meiotic pairing by investigation of different alleles of genes subject to MSUD at various locations in the genome. The mechanism of meiotic silencing—involvement of RNA RT-PCR analysis of sad-1 transcripts revealed expression is specific to sexual tissue and, furthermore, no induction by the presence of unpaired DNA was observed in the crosses used for RNA isolation.(2) sad-1 encodes a protein with significant similarity to RNA-dependent RNA polymerases involved in silencing in a variety of organisms, including QDE-1, a protein essential for quelling in N. crassa.(2,19) Therefore, one may conclude that synthesis of double stranded RNA and probably RNA amplification is involved in MSUD, a step in common with quelling and other PTGS mechanisms. It has become clear that PTGS involves sensing aberrant RNA, formation of dsRNA which is a substrate for the Dicer ribonuclease to generate siRNAs which guide destruction of homologous mRNA via a protein complex (RNA-induced silencing complex- RISC) containing another nuclease and are also used to prime RdRP amplification to generate more siRNA via DICER.(5,10,20) siRNA acts in trans to silence transcripts and can be propagated between cells through vegetative tissue.(9,21–23) MSUD has been shown, using mosaic perithecia arising from crosses involving a heterokaryotic female, to be restricted to the ascus containing unpaired DNA.(2) The story, to date, provokes several interesting questions. For instance, how is the upstream signal for RNA-dependent silencing detected? For quelling, N. crassa is able to detect additional copies of normal genes. The signal for MSUD must be unpaired meiotic DNA which could lead to the formation of transcripts not normally synthesised because of pairing. Is any unpaired region of a gene sufficient to trigger MSUD or is either the 50 or 30 necessary? What other genes are involved in MSUD? It seems likely that other components of the MSUD BioEssays 25.2 101 What the papers say pathway will be conserved with the PTGS machinery. Inspection of the N. crassa genome (Neurospora Sequencing Project. Assembly version 3. Whitehead Institute/MIT Center for Genome Research (http://www-genome.wi.mit.edu)) indicates the presence of dicer orthologs. Is one of these specific to sexual development? Steps in the MSUD pathway must play a normal essential role in meiosis since crosses homozygous for Sad-1 mutations are barren.(2) Therefore other MSUD proteins may be involved in meiosis-perhaps a DICER function generating one or more essential stRNAs. Genetic dissection of MSUD via a comprehensive isolation of additional Sad genes will indicate whether MSUD and quelling use common components and provide a unique opportunity for analysing the requirements for pairing and its role in determining the progression of the meiotic cell cycle. General implications Silencing mechanisms identified in N. crassa are likely to play a significant role in controlling invasion of the genome by transposable elements and viruses.(24) RIP causes permanent gene inactivation and is probably of greatest importance in resisting such invasions, as has been shown by studies of the Tad-1 element in various N. crassa strains.(25) Quelling will silence expression of transposon sequences present in multiple copies restricting transposition in vegetative cells. MSUD now adds a further mechanism for maintenance of genome integrity as a cross between a strain containing one or more transposons and one lacking such sequences will result in unpaired meiotic DNA allowing MSUD to silence genes required for transposition at perhaps a very vulnerable stage for genome integration. The effects of Sad mutations on transposition frequencies will test this proposal. Just within the fungi interesting questions arise. Many fungi have no known sexual cycle—the so-called Imperfect Fungi— and it has been a long standing question whether this is because the correct strains or conditions for detection of mating have not been discovered or whether, as indicated by population and evolutionary studies,(26–28) sexual reproduction has been lost. The genome of Aspergillus fumigatus, a species lacking a known sexual phase, contains clear orthologs of both qde-1 and sad-1 (The Institute for Genomic Research, http:// www.tigr.org ). Since sad-1 expression is restricted to meiosis this indicates that sex may yet be discovered in this species. A less likely scenario is that in some species of fungi sad-1 may play a role in vegetative cells. As more fungal genome sequences become available the extent of conservation of silencing mechanisms will be of great interest. The essential role of the MSUD pathway in meiosis raises important questions regarding its generality. The special advantages of Neurospora for analysis of individual meioses permitted its discovery in the first place. Finding orthologs of Sad genes in other species will allow their study in normal meiotic processes as well as in the context of interspecies crosses. 102 BioEssays 25.2 Acknowledgments We thank David Perkins for useful comments on the manuscript. Neurospora crassa genome sequence data was obtained from the Neurospora Sequencing Project, assembly version 3, Whitehead Institute/MIT Center for Genome Research (http://www-genome.wi.mit.edu). Preliminary Aspergillus fumigatus sequence data was obtained from The Institute for Genomic Research website (http://www. tigr.org). References 1. Shiu PK, Raju NB, Zickler D, Metzenberg RL. Meiotic silencing by unpaired DNA. Cell 2001;107:905–916. 2. Shiu PK, Metzenberg RL. Meiotic Silencing by Unpaired DNA. Properties, regulation and suppression. Genetics 2002;161:1483–1495. 3. Boutla A, Delidakis C, Livadaras I, Tsagris M, Tabler M. Short 50 phosphorylated double-stranded RNAs induce RNA interference in Drosophila. Curr Biol 2001;11:1776–1780. 4. Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 2001;293:2269–2271. 5. Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001; 409:363–366. 6. Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001;293: 834–838. 7. Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 2001;15: 2654–2659. 8. Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 2001;106:23–34. 9. Golden TA, Schauer SE, Lang JD, Pien S, Mushegian AR, Grossniklaus U, Meinke DW, Ray A. Short Integuments1/suspensor1/carpel Factory, a Dicer Homolog, Is a Maternal Effect Gene Required for Embryo Development in Arabidopsis. Plant Physiol 2002;130:808–822. 10. Cogoni C. Homology-dependent gene silencing mechanisms in fungi. Annu Rev Microbiol 2001;55:381–406. 11. Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNAdependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 2000;101:543–553. 12. Selker EU. Repeat-induced gene silencing in fungi. Adv Genet 2002;46: 439–450. 13. Raju NB. Genetic control of the sexual cycle in Neurospora. Mycol Res 1992;96:241–262. 14. Glass NL, Vollmer SJ, Staben C, Grotelueschen J, Metzenberg RL, Yanofsky C. DNAs of the two mating-type alleles of Neurospora crassa are highly dissimilar. Science 1988;241:570–573. 15. Pickford AS, Catalanotto C, Cogoni C, Macino G. Quelling in Neurospora crassa. Adv Genet 2002;46:277–303. 16. Cogoni C, Macino G. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc Natl Acad Sci USA 1997;94:10233–10238. 17. Catalanotto C, Azzalin G, Macino G, Cogoni C. Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev 2002;16:790–795. 18. Aramayo R, Metzenberg RL. Meiotic transvection in fungi. Cell 1996;86: 103–13. 19. Cogoni C, Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 1999; 399:166–169. What the papers say 20. Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 2001;293:1146–1150. 21. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998;391:806–811. 22. Voinnet O, Baulcombe DC. Systemic signalling in gene silencing. Nature 1997;389:553. 23. Palauqui JC, Elmayan T, Pollien JM, Vaucheret H. Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J 1997; 16:4738–4745. 24. Plasterk RH. RNA silencing: the genome’s immune system. Science 2002;296:1263–1265. 25. Kinsey JA, Garrett-Engele PW, Cambareri EB, Selker EU. The Neurospora transposon Tad is sensitive to repeat-induced point mutation (RIP). Genetics 1994;138:657–664. 26. Geiser DM, Arnold ML, Timberlake WE. Sexual origins of British Aspergillus nidulans isolates. Proc Natl Acad Sci USA 1994;91:2349–2352. 27. Geiser DM, Timberlake WE, Arnold ML. Loss of meiosis in Aspergillus. Mol Biol Evol 1996;13:809–817. 28. Taylor J, Jacobson D, Fisher M. The evolution of asexual fungi: Reproduction, Speciation and Classification. Annu Rev Phytopathol 1999;37:197–246. BioEssays 25.2 103