* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Types of RNA
Gene expression profiling wikipedia , lookup
Alternative splicing wikipedia , lookup
Gel electrophoresis wikipedia , lookup
Biochemistry wikipedia , lookup
Bottromycin wikipedia , lookup
X-inactivation wikipedia , lookup
Molecular evolution wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Non-coding DNA wikipedia , lookup
Gene regulatory network wikipedia , lookup
List of types of proteins wikipedia , lookup
Genetic code wikipedia , lookup
Biosynthesis wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
RNA interference wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Polyadenylation wikipedia , lookup
Messenger RNA wikipedia , lookup
Gene expression wikipedia , lookup
RNA silencing wikipedia , lookup
Piccole molecole di RNA: i microRNAs e la loro ruolo funzionale
https://en.wikipedia.org/wiki/MicroRNA#/media/File:MiRNA.svg
Interaction of microRNAs and mRNA 3’ UTR region
miRNA-mRNA interaction model. a. miRNA binding site in loop region of mRNA (favorable); b. miRNA
binding site in stem region of mRNA (unfavorable), c. miRNA binding site in stem-loop junction (sequence
dependent).
http://openi.nlm.nih.gov/detailedresult.php?img=2676258_1471-2105-10-108-1&req=4
Interaction of microRNAs with different alles causing mRNA binding
'U' allele
'C' allele
The validated binding site for hsa-miR-24 in the 3' UTR of DHFR (dihydrofolate reductase) gene with 'U' allele
14 bp downstream is structured and hence, inaccessible for miRNA binding, while the 'C' allele makes the
target site totally unstructured thereby allowing miRNA binding.
http://openi.nlm.nih.gov/detailedresult.php?img=2676258_1471-2105-10-108-1&req=4
Prediction of 3’UTR structure of target mRNA and miR-binding
Prediction of target mRNA and miR-binding
microRNA processing
https://www.youtube.com/watch?v=xRoenh-LERY
microRNAs used to modulate gene expression
Purification small RNAs from samples
such as serum, plasma, urine and cerebrospinal fluid
RNA Isolation kits are high quality kits optimized for the purification of small RNAs
from serum, plasma and other biofluids. Based on spin column chromatography, the
kit uses a proprietary resin as the separation matrix (without the use of toxic phenol
or chloroform).
Use the kit to isolate all RNAs smaller than 1000 nt, from mRNA and tRNA down to
microRNA and small interfering RNA (siRNA), from other cell components, such as
proteins.
Take advantage of the optimized yet fast and easy-to-use protocols based on either
centrifugation or a vacuum manifold.
RNA isolated using these kits are ideal for highly sensitive quantification of
microRNA in applications such as biomarker discovery and toxicology
studies. It is also well suited for other downstream applications, including miRprofiling.
Applications: Biomarker identification
Purification of mRNA targets of miRNA
Lo psoralene è il capostipite di una famiglia di
composti organici naturali, le furanocumarine lineari,
derivati dalla cumarina mediante l'aggiunta di un anello
furanico e può essere considerato come un derivato
dell'umbelliferone.
Lo psoralene è presente in natura in alcune piante.
miR
synthetic miR
mRNA
Andrew Grimson. Nature Chemical Biology 11, 100–101 (2015)
Per effetto dei raggi UV, che determinano un aumento
della sua reattività, lo psoralene è in grado di legarsi a
ponte fra due timine. Questo meccanismo è sfruttato
in medicina per curare malattie come la psoriasi,
eczemi,
vitiligine.
Tale
tecnica
è
detta
fotochemioterapia ed è usata anche per la cura di altre
patologie cutanee.
Il miR-CLIP utilizza un miRNA sintetico per
catturare l'RNA bersaglio. Una molecola di
psoralene è legato al miR e lo lega
covalentemente all’mRNA bersaglio.
Il miR è anche legato ad una molecola di
biotina, che facilita la purificazione per
immunoprecipitazione
con
anticorpi
associati a streptavidina. Gli mRNA sono poi
sequenziati per deep sequencing, senza
staccarli dal miR.
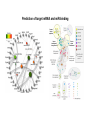
Potential mechanisms of miRNAmediated downregulation.
Ribonucleoproteins associate ai
miRNA (miRNPs) target mRNAs
through sequence
complementation. The association
between miRNPs and mRNAs
can have one or several
consequences. miRNP-mediated
downregulation may have direct
and indirect mechanisms and
occur before translation is
triggered or after translation
initiation.
hnRNA (heterogeneous nuclear
RNA) is a group of molecules which
includes the pre-mRNA transcripts
and other complexed proteins forming
the hnRNP (heterogeneous nuclear
ribonucleoprotein particles), not
destined to become cytoplasmic
mRNA. Despite this distinction the
hnRNAs is often used synonymously
with the pre-mRNA.
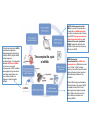
The complex life cycle
of mRNA
mRNP (messenger
ribonucleoprotein) is mRNA with
bound proteins. mRNA does not exist
in vivo "naked", mRNA is always
bound by many different proteins while
being synthesized, spliced, exported
and while being translated in the
cytoplasm.
When mRNA is being synthesized by
RNA polymerase, this nascent mRNA
is already bound by RNA 5′ end 7methyl-guanosine capping enzymes.
Later mRNA is bound by exon and
intron definition complexes, and
splicing snRNPs.
mRNA processing
Modification of hnRNA (heterogeneous
nuclear RNA):
1. Capping addition of 7-methyl
Guanosine at the 5’ end
2. Poly-A tail added to the 3’ end
3. Introns are removed by a process
called splicing to produce the mature
mRNA
mRNA processing
Mechanisms of RNA
transcript modification
Nonsense Mediated
mRNA Decay (NMD):
detection and decay of
mRNA transcripts
containing premature
termination codons
Nonstop Mediated mRNA
Decay (NSD): detection
and decay of mRNA
transcripts which lack a
stop codon
Mechanisms of control
of the levels of protein
synthesized from its
mRNA.
Types of RNA
•
Messenger RNA (mRNA) is the RNA that carries information
from DNA to the ribosome, the sites of protein synthesis
(translation) in the cell. The coding sequence of the mRNA
determines the amino acid sequence in the protein that is
produced. However, many RNAs do not code for protein
(about 96% of the transcriptional output is non-protein-coding
in eukaryotes).
•
These so-called non-coding RNAs ("ncRNA") can be
encoded by their own genes (RNA genes), but can also
derive from mRNA introns. The most prominent examples of
non-coding RNAs are transfer RNA (tRNA) and ribosomal
RNA (rRNA), both of which are involved in the process of
translation. There are also non-coding RNAs involved in gene
regulation, RNA processing and other roles. Certain RNAs
are able to catalyse chemical reactions such as cutting and
ligating other RNA molecules, and the catalysis of peptide
bond formation in the ribosome; these are known as
ribozymes.
Types of RNA: in translation
Messenger RNA (mRNA) carries information about a protein sequence to the ribosomes, the protein synthesis factories in the cell. It is
coded so that every three nucleotides (a codon) correspond o one amino acid. In eukaryotic cells, once precursor mRNA (pre-mRNA) has
been transcribed from DNA, it is processed to mature mRNA. This removes its introns—non-coding sections of the pre-mRNA. The mRNA
is then exported from the nucleus to the cytoplasm, where it is bound to ribosomes and translated into its corresponding protein form with
the help of tRNA. In prokaryotic cells, which do not have nucleus and cytoplasm compartments, mRNA can bind to ribosomes while it is
being transcribed from DNA. After a certain amount of time the message degrades into its component nucleotides with the assistance of
ribonucleases.
Transfer RNA (tRNA) is a small RNA chain of about 80 nucleotides that transfers a specific amino acid to a growing polypeptide chain at
the ribosomal site of protein synthesis during translation. It has sites for amino acid attachment and an anticodon region for codon
recognition that binds to a specific sequence on the messenger RNA chain through hydrogen bonding.
Ribosomal RNA (rRNA) is the catalytic component of the ribosomes. Eukaryotic ribosomes contain four different rRNA molecules: 18S,
5.8S, 28S and 5S rRNA. Three of the rRNA molecules are synthesized in the nucleolus, and one is synthesized elsewhere. In the
cytoplasm, ribosomal RNA and protein combine to form a nucleoprotein called a ribosome. The ribosome binds mRNA and carries out
protein synthesis. Several ribosomes may be attached to a single mRNA at any time. Nearly all the RNA found in a typical eukaryotic cell is
rRNA.
Transfer-messenger RNA (tmRNA) is found in many bacteria and plastids. It tags proteins encoded by mRNAs that lack stop codons for
degradation and prevents the ribosome from stalling.
Types of RNA: regulatory RNAs
Several types of RNA can downregulate gene expression by being complementary to a part of an mRNA or a gene's DNA. MicroRNAs
(miRNA; 21-22 nt) are found in eukaryotes and act through RNA interference (RNAi), where an effector complex of miRNA and enzymes
can cleave complementary mRNA, block the mRNA from being translated, or accelerate its degradation.
While small interfering RNAs (siRNA; 20-25 nt) are often produced by breakdown of viral RNA, there are also endogenous sources of
siRNAs. siRNAs act through RNA interference in a fashion similar to miRNAs. Some miRNAs and siRNAs can cause genes they target to
be methylated, thereby decreasing or increasing transcription of those genes. Animals have Piwi-interacting RNAs (piRNA; 29-30 nt) that
are active in germline cells and are thought to be a defense against transposons and play a role in gametogenesis.
Many prokaryotes have CRISPR RNAs, a regulatory system similar to RNA interference. Antisense RNAs are widespread; most
downregulate a gene, but a few are activators of transcription. One way antisense RNA can act is by binding to an mRNA, forming doublestranded RNA that is enzymatically degraded. There are many long noncoding RNAs that regulate genes in eukaryotes, one such RNA is
Xist, which coats one X chromosome in female mammals and inactivates it.
Types of RNA: in RNA processing
Many RNAs are involved in modifying other RNAs. Introns are spliced out of pre-mRNA by spliceosomes, which contain several small
nuclear RNAs (snRNA), or the introns can be ribozymes that are spliced by themselves. RNA can also be altered by having its
nucleotides modified to other nucleotides than A, C, G and U. In eukaryotes, modifications of RNA nucleotides are in general directed by
small nucleolar RNAs (snoRNA; 60-300 nt), found in the nucleolus and cajal bodies. snoRNAs associate with enzymes and guide them to
a spot on an RNA by basepairing to that RNA. These enzymes then perform the nucleotide modification. rRNAs and tRNAs are extensively
modified, but snRNAs and mRNAs can also be the target of base modification. RNA can also be methylated.
Types of RNA: RNA genomes
Like DNA, RNA can carry genetic information. RNA viruses have genomes composed of RNA that encodes a number of
proteins. The viral genome is replicated by some of those proteins, while other proteins protect the genome as the virus
particle moves to a new host cell. Viroids are another group of pathogens, but they consist only of RNA, do not encode
any protein and are replicated by a host plant cell's polymerase.
Reverse transcribing viruses replicate their genomes by reverse transcribing DNA copies from their RNA; these DNA
copies are then transcribed to new RNA. Retrotransposons also spread by copying DNA and RNA from one another, and
telomerase contains an RNA that is used as template for building the ends of eukaryotic chromosomes.
Double-stranded RNA (dsRNA) is RNA with two complementary strands, similar to the DNA found in all cells. dsRNA
forms the genetic material of some viruses (double-stranded RNA viruses). Double-stranded RNA such as viral RNA or
siRNA can trigger RNA interference in eukaryotes, as well as interferon response in vertebrates.
How much RNA does a typical
mammalian cell contain?
The RNA content and RNA make up of a cell depend very much on its
developmental stage and the type of cell.
To estimate the approximate yield of RNA that can be expected from your starting
material, we usually calculate that a typical mammalian cell contains 10–30 pg
total RNA.
The majority of RNA molecules are tRNAs and rRNAs.
mRNA accounts for only 1–4% of the total cellular RNA although the actual
amount depends on the cell type and physiological state. Approximately 360.000
mRNA molecules are present in a single mammalian cell, made up of
approximately 12.000 different transcripts with a typical length of around 2 kb.
Some mRNAs comprise 3% of the mRNA pool whereas others account for
less than 0.1%. These rare or low-abundance mRNAs may have a copy number
of only 5–15 molecules per cell.
(PTM)
Northern blot
Developed by James Alwine & George Stark, at Stanford University in 1977
Northern Blot is a technique for detection of specific RNA sequences
Load 10 µgr of total RNA
RNA molecules have defined length and much
shorter than genomic DNA, so it is not necessary to
cleave RNA before electrophoresis
RNA are more sensible to degradation than DNA
RNA molecules are separated based on size by gel
electrophoresis
RNA presents secondary structure and need to be
denatured in formaldehyde agarose gel
RNA is immobilized by heat
or UV light
hybridization buffer: 50%
formamide, 0.5% SDS,
5xSSPE, 5x Denhardt’s
solution, and 20 μg/mL
sheared, denatured, salmon
sperm DNA
washed at low stringency
in 2xSSC, 0.1% SDS at
34-45°C twice for five
minutes or at high
stringency in 0.1 SSC,
0.1% SDS at 65°C twice
for five minutes
The bands obtained by
autoradiography give a
measure of the amount and
size of specific mRNA in the
sample
Techniques to study mRNA and
microRNA expression
miRNA research tools
miRNA-profiling chips
1100
http://www.microrna.org/microrna/home.do
717
387
miRNA-profiling
Ogni campione viene marcato
con una specifica molecola
fluorescente (Cyanine è un
nome non specific per questa
famiglia di coloranti sintetici )
Chip con legate tutte le
sonde per tutti i geni murini
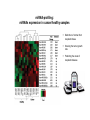
miRNA-profiling:
miRNAs expression in cancer/healthy samples
Distinction of normal from
neoplastic tissue
Knowing the tumor growth
rate
Predicting the onset of
neoplastic disease
Individuazione di mRNA modulati da microRNA