* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download compgenomics
Quantitative trait locus wikipedia , lookup
Gene nomenclature wikipedia , lookup
DNA sequencing wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Gene desert wikipedia , lookup
Genomic imprinting wikipedia , lookup
Frameshift mutation wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
History of genetic engineering wikipedia , lookup
Public health genomics wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Non-coding RNA wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Human genome wikipedia , lookup
Genome (book) wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Gene expression programming wikipedia , lookup
Epigenomics wikipedia , lookup
Non-coding DNA wikipedia , lookup
Minimal genome wikipedia , lookup
Whole genome sequencing wikipedia , lookup
Oncogenomics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Designer baby wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Point mutation wikipedia , lookup
Genomic library wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Gene expression profiling wikipedia , lookup
Genome evolution wikipedia , lookup
Pathogenomics wikipedia , lookup
Microevolution wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Helitron (biology) wikipedia , lookup
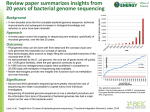
Igor Ulitsky “the branch of genetics that studies organisms in terms of their genomes (their full DNA sequences)” Computational genomics in TAU ◦ Ron Shamir’s lab – focus on gene expression and regulatory networks ◦ Eithan Ruppin’s lab – focus on metabolism ◦ Tal Pupko’s and Benny Chor’s labs – focus on phylogeny ◦ Roded Sharan’s lab – focus on networks ◦ Noam Shomron’s lab – focus on miRNA ◦ Eran Halperin’s lab – focus on genetics Alignment Protein coding gene finding Assembly of long reads Basic microarray data analysis Mapping of transcriptional regulation in simple organisms Functional profiling in simple organisms Determining protein abundance Assembly of short reads Transcriptional regulation in higher eukaryotes “Histone code”: Chromatin modifications, their function and regulation Functional profiling of mammalian cells Association studies for single-gene effects Construction and modeling of synthetic circuits Digital gene expression from RNA-seq studies Prediction of ncRNAs and their function Global mapping of alternative splicing regulation Integration of multi-level signaling (TFs, miRNA, chromatin) Association studies for combinations of alleles All microbial genomes are sequenced in E. coli Each sequencing efforts basically introduces genes (3-8Kb fragments) into E. coli Sometimes sequencing fails Idea: sequencing fails barrier to horizontal gene transfer Even sequencing of reads with 100s of bp will no identify many indels Idea: sequence pairs of sequences at some distance apart from each other High-throughput sequencing can identify all the mutations in different cancers 20,857 transcripts from 18,191 human genes sequenced in 11 breast and 11 colorectal cancers. Problems: few mutations are drivers most are passangers Most studies did not identify high frequent risk allels But: members of some pathways are affected in almost any tumour Network biology needed Using histone modifications and sequence conservation to uncover long noncoding RNAs (lincRNA) 12 fly species were sequenced to identify ◦ Evolution of genes and chromosome ◦ Evolutionary constrained sequence elements in promoters and 3’ UTRs Starting point – genome-wide alignment of the genomes