* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Supplementary Information (doc 33K)
Expanded genetic code wikipedia , lookup
Molecular evolution wikipedia , lookup
Protein moonlighting wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Gel electrophoresis wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Gene expression wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Protein adsorption wikipedia , lookup
Protein–protein interaction wikipedia , lookup
List of types of proteins wikipedia , lookup
Bottromycin wikipedia , lookup
Community fingerprinting wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
RNA interference wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Genetic code wikipedia , lookup
Western blot wikipedia , lookup
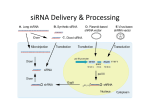
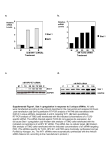
Supplementary Materials and Methods Mass spectrometry Immunoprecipitates were separated by SDS-PAGE and stained with silver. Specific silver-stained gel bands were excised, destained, dehydrated, and alkylated. After gel pieces were dried, proteins were digested with trypsin and peptides were extracted. Each tryptic peptide sample was dissolved and analyzed by LTQ (Thermo). MS data were searched using SEQUEST algorithm against the National Center for Biotechnology Information (NCBI) human protein database and results were filtered, sorted, and displayed using the Bioworks 3.2 softerware. Construction of recombinant Lentivirus vectors with shRNA for hCINAP The siRNA sequence for hCINAP (5’-CAGAGUAGUUGAUGAGUUA-3’) was selected after screening to validate potential siRNAs (25). Non-silencing siRNA (5’-UUCUCCGAACGUGUCACGU-3’) was used as a control. Corresponding to siRNA sequences for hCINAP and non-silencing, shRNAs were synthesized and inserted into downstream of the U6 promoter in the lentiviral vector pGCSIL-Puromycin (GeneChem Co., Ltd., Shanghai, China). To further verify that the effects of hCINAP RNAi are specific, we prepared two constructs bearing two and three-point mutations in the third base of codons within the 19-bp RNAi sequence (5’-CAGA180GUA183GTT186GATGAGTTA-3’) targeting hCINAP expression. The silent mutations in the third base of codon didn’t change the amino acid sequence of hCINAP protein but rendered its expression insensitive to inhibition by the shRNA. The RNAi-resistant mutants were named as hCINAP mutant 1 (A180G, A183C), hCINAP mutant 2 (A180G, A183C, T186C). Ribosome profiling At 48 h after transfection with indicated plasmids, HeLa cells were treated with 100 μg/ml Cycloheximide (CHX) for 10 min. Cells were then washed with pre-cold PBS (containing 100 μg/ml CHX) and resuspended in hypotonic buffer (10 mM HEPES pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 100 μg/ml CHX, 40 U/ml RNase Inhibitor, 1 x Cock tail) on ice for 15 min. The cells were mechanically disrupted by 15 strokes with Dounce homogenizer (KONTES). The homogenate was centrifuged at 720 g for 10 min, and the supernatant was collected and considered as the cytoplasmic fraction. 1 mg of total proteins was loaded onto a linear sucrose gradient (5-50%) prepared with a Gradient Master former (BioComp Instruments). The samples were centrifuged for 150 min at 36,000 rpm at 4°C in a SW41 rotor (Beckman). The gradient was then analyzed at OD254 nm with BioComp Gradient Fractionator to get ribosome profiles.