* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Slide 1
Molecular cloning wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Genetic code wikipedia , lookup
Genome evolution wikipedia , lookup
List of types of proteins wikipedia , lookup
Community fingerprinting wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Gene expression profiling wikipedia , lookup
Gene regulatory network wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Biosynthesis wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Non-coding DNA wikipedia , lookup
Molecular evolution wikipedia , lookup
Polyadenylation wikipedia , lookup
Point mutation wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Non-coding RNA wikipedia , lookup
RNA silencing wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Messenger RNA wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Gene expression wikipedia , lookup
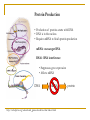
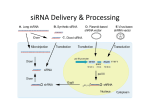
New approaches for determining functional siRNA Liyang Diao Dr. Stanley Dunn, advisor Protein Production • Production of proteins starts with DNA • DNA is in the nucleus • Requires mRNA to finish protein production mRNA: messenger RNA RNAi: RNA interference • Suppresses gene expression • Affects mRNA DNA mRNA http://nobelprize.org/educational_games/medicine/dna/index.html protein More on RNAi siRNA: short-interfering RNA • Typically 20-25 nucleotides long • Double-stranded • Participates in RNAi by degrading mRNA Potential for effective gene therapy Issues • Some genes are more effectively suppressed than others • Mechanism is poorly understood Diagram: http://www.ambion.com/techlib/append/RNAi_mechanism.html Question How do we know which siRNA are functional? Some ideal properties: GC content between 30-55% Low level of secondary structure Differential between thermodynamic stability of 5’ and 3’ ends: A/U content Specific positional nucleotide preferences Avoid long GC stretches http://bioinf.man.ac.uk/resources/phase/manual/RNAMolecule.png Previous Model Pancoska’s Eulerian graph model • Represent a string of siRNA by a directed digraph first • Construct a weighted undirected Eulerian graph A T G C • Compare graphs for functional and non functional siRNA • For these two sets of siRNA, compute graph properties that reflect sequence structure. Issues with Pancoska’s Algorithm A T G C ATTCGTGGACG GATTCGTGGAC CGATTCGTGGA … • Uniqueness • Complex pattern recognition Other Ideas • Number of nucleotide mutations • Levenshtein distance: Measures the minimum number of substitutions/insertions required to go from one string to another. Current/Future Progress • 420 total number of possible siRNA strands of length 20. • How many are potentially functional? • Combinatorics! Math • Let H(n,i,j) be the number of potential positions of A/U, G/C pairs. • • • • Thus, the total number of potential strings is 220 * H(n,i,j). n the total number of G or C nucleotides i the total number of A or U nucleotides at 5’ end j the total number of A or U nucleotides at 3’ end Quantity desired: