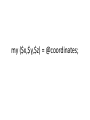* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Survey
Document related concepts
Transcript
Perl
Introduction
Why Perl?
•
•
•
•
•
•
Widely used scripting language
Powerful text manipulation capabilities
Relatively easy to use
Has a wide range of libraries available
Fast
Good support for file and process operations
Less suiteable for:
• Building large and complex applications
– Java, C\C++, C#
• Applications with a GUI
– Java, C\C++, C#
• High performance/memory efficient applications
– Java, C\C++, C#, Fortran
• Statistics
–R
Learning to script
Knowledge +
Skills
Exercise
Determine the percentage GC-content of the
human chromosome 22
open file
read lines
per line:
skip if header line
count Cs and Gs
count all nucleotides
report percentage Cs and Gs
Hello World
Hello World….
Simple line of Perl code:
print "Hello World";
Run from a terminal:
perl -e 'print "Hello World";'
Now try this and notice the difference:
perl -e 'print "Hello World\n";'
\n
“backslash-n”
newline character
'Enter'key
\t
“backslash-t”
'Tab' key
Hello World (cont)
To create a text file with this line of Perl code:
echo 'print "Hello World\n";' > HelloWorld.pl
perl HelloWorld.pl
In the terminal window, type
kate HelloWorld.pl
and then hit the enter key. Now you can edit the Perl code.
Pythagoras' theorem
a2 + b 2 = c 2
32 + 42 = 52
Pythagoras.pl
$a =
$b =
$a2 =
$b2 =
$c2 =
$c =
print
3;
4;
$a * $a;
$b * $b;
$a2 + $b2;
sqrt($c2);
$c;
$a
a single value or scalar variable
starts with a $ followed by its name
Pythagoras.pl
$a =
$b =
$a2 =
$b2 =
$c2 =
$c =
print
3;
4;
$a * $a;
$b * $b;
$a2 + $b2;
sqrt($c2);
$c;
5
Perl scripts
Add these lines at the top of each Perl script:
#!/usr/bin/perl
# author:
# description:
use strict;
use warnings;
perl Pythagoras.pl
Global symbol "$a2" requires explicit package name at Pythagoras.pl line 8.
Global symbol "$b2" requires explicit package name at Pythagoras.pl line 9.
Global symbol "$c2" requires explicit package name at Pythagoras.pl line 10.
Global symbol "$a2" requires explicit package name at Pythagoras.pl line 10.
Global symbol "$b2" requires explicit package name at Pythagoras.pl line 10.
Global symbol "$c" requires explicit package name at Pythagoras.pl line 11.
Global symbol "$c2" requires explicit package name at Pythagoras.pl line 11.
Global symbol "$c" requires explicit package name at Pythagoras.pl line 12.
Execution of Pythagoras.pl aborted due to compilation errors.
Pythagoras.pl
$a =
$b =
$a2 =
$b2 =
$c2 =
$c =
print
3;
4;
$a * $a;
$b * $b;
$a2 + $b2;
sqrt($c2);
$c;
Pythagoras.pl
my $a = 3;
my $b = 4;
my $a2 = $a * $a;
my $b2 = $b * $b;
my $c2 = $a2 + $b2;
my $c = sqrt($c2);
print $c;
my
The first time a variable appears in
the script, it should be claimed using
‘my’. Only the first time...
Pythagoras.pl
my($a,$b,$c,$a2,$b2,$c2);
$a = 3;
$b = 4;
$a2 = $a * $a;
$b2 = $b * $b;
$c2 = $a2 + $b2;
$c = sqrt($c2);
print $c;
Pythagoras.pl
$a =
$b =
$a2 =
$b2 =
$c2 =
$c =
print
3;
4;
$a * $a;
$b * $b;
$a3 + $b2;
sqrt($c2);
$c;
4
Pythagoras.pl
$a =
$b =
$a2 =
$b2 =
$c2 =
$c =
print
3;
4;
$a * $a;
$b * $b;
$a3 + $b2;
sqrt($c2);
$c;
Pythagoras.pl
my $a = 3;
my $b = 4;
my $a2 = $a * $a;
my $b2 = $b * $b;
my $c2 = $a3 + $b2;
my $c = sqrt($c2);
print $c;
perl Pythagoras.pl
Global symbol "$a3" requires explicit package name at Pythagoras.pl line 10.
Execution of Pythagoras.pl aborted due to compilation errors.
Text or number
Variables can contain text (strings) or numbers
my $var1 = 1;
my $var2 = "2";
my $var3 = "three";
Try these four statements:
print $var1 + $var2;
print $var2 + $var3;
print $var1.$var2;
print $var2.$var3;
Text or number
Variables can contain text (strings) or numbers
my $var1 = 1;
my $var2 = "2";
my $var3 = "three";
Try these four statements:
print $var1 + $var2;
print $var2 + $var3;
print $var1.$var2;
print $var2.$var3;
=>
=>
=>
=>
3
2
12
2three
variables can be added, subtracted, multiplied,
divided and modulo’d with:
+
-
*
/
%
variables can be concatenated with:
.
sequence.pl
print "Please type a DNA sequence: ";
#this is a comment line
#Read a line from the standard input (keyboard)
my $DNAseq = <STDIN>;
#Remove the newline (Enter) from the typed text
chomp($DNAseq);
#Get the length of the text(DNA sequence)
my $length = length($DNAseq);
print "It has $length nucleotides\n";
Program flow
is top - down
sequence.pl
print "Please type a DNA sequence: ";
#this is a comment line
#Read a line from the standard input (keyboard)
my $DNAseq = <STDIN>;
#Remove the newline (Enter) from the typed text
chomp($DNAseq);
#Get the length of the text(DNA sequence)
my $length = length($DNAseq);
print "It has $length nucleotides\n";
<STDIN>
read characters that are typed on the
keyboard. Stop after the Enter key is
pressed
<>
same, STDIN is the default and can be
left out. This is a recurring and
confusing theme in Perl...
sequence.pl
print "Please type a DNA sequence: ";
#this is a comment line
#Read a line from the standard input (keyboard)
my $DNAseq = <>;
#Remove the newline (Enter) from the typed text
chomp($DNAseq);
#Get the length of the text(DNA sequence)
my $length = length($DNAseq);
print "It has $length nucleotides\n";
$output = function($input)
input and output can be left out
parentheses are optional
$coffee = function($beans,$water)
sequence2.pl
print "Please type a DNA sequence: ";
my $DNAseq = <>;
chomp($DNAseq);
#Get the first three characters of $DNAseq
my $first3bases = substr($DNAseq,0,3);
print "The first 3 bases: $first3bases\n";
$frag = substr($text, $start, $num)
Extract a fragment of string $text starting
at $start and with $num characters.
The first letter is at position 0!
perldoc
perldoc -f substr
substr EXPR,OFFSET,LENGTH,REPLACEMENT
substr EXPR,OFFSET,LENGTH
substr EXPR,OFFSET
Extracts a substring out of EXPR and
returns it.
First character is at offset 0, .....
print
perldoc -f print
print FILEHANDLE LIST
print LIST
print Prints a string or a list of strings.
If you leave out the FILEHANDLE, STDOUT is the
destination: your terminal window.
print
In Perl items in a list are separated by commas
print "Hello World","\n";
Is the same as:
print "Hello World\n";
sequence3.pl
print "Please type a DNA sequence: ";
my $DNAseq = <>;
chomp($DNAseq);
#Get the second codon of $DNAseq
my $codon2 = substr($DNAseq,3,3);
print "The second codon: $codon2\n";
if, else, unless
sequence4.pl
print "Please type a DNA sequence: ";
my $DNAseq = <>;
chomp($DNAseq);
#Get the first three characters of $DNAseq
my $codon = substr($DNAseq,0,3);
if($codon eq "ATG") {
print "Found a start codon\n";
}
Conditional execution
if ( condition ) {
do something
}
if ( condition ) {
do something
} else {
do something else
}
Conditional execution
if ( $number > 10 ) {
print "larger than 10";
} elsif ( $number < 10 ) {
print "smaller less than 10";
} else {
print "number equals 10";
}
unless ( $door eq "locked" ) {
openDoor();
}
Conditions are true or false
1 < 10 : true
21 < 10 : false
Comparison operators
Numeric test
String test
==
!=
>
>=
eq
ne
gt
ge
<
<=
<=>
lt
le
cmp
Meaning
Equal to
Not equal to
Greater than
Greater than or equal to
Less than
Less than or equal to
Compare
Examples
if
if
if
if
if
if
if
(
(
(
(
(
(
(
1 ==
1 ==
1 !=
-1 >
"hi"
"hi"
"hi"
1 ) { # TRUE
2 ) { # FALSE
2 ) { # TRUE
10 ) { # FALSE
eq "dag" ) { # FALSE
gt "dag" ) { # TRUE
== "dag" ) { # TRUE !!!
The last example may surprise you, as "hi" is not equal to "dag"
and therefore should evaluate to FALSE. But for a numerical
comparison they are both 0.
numbers as conditions
0 : false
all other numbers : true
Numbers as conditions
if ( 1 ) {
print "1 is true";
}
if ( 0 ) {
print "this code will not be reached";
}
if ( $open ) {
print "open is not zero";
}
repetition
sequence5.pl
print "Please type a DNA sequence: ";
my $DNAseq = <>;
chomp($DNAseq);
#Get all codons of $DNAseq
my $position = 0
while($position < length($DNAseq)) {
my $codon = substr($DNAseq,$position,3);
print "The next codon: $codon\n";
$position = $position + 3;
}
the while loop
while ( condition ) {
do stuff
}
my $i = 0;
while ($i < 10) {
$i = $i + 1;
}
print $i;
$i = $i + 1
First the part to the right of the
assignment operator ‘=‘ is
calculated, then the result is moved
to the left.
$i += 1
Same result as previous slide.
$i++
Same as result previous slide,
increments $i with 1.
++$i
Same as previous, but compare:
print $i++;
print ++$i;
Exercise: Fibonacci numbers
Write a script that calculates and prints all
Fibonacci numbers below one thousand.
0, 1, 1, 2, 3, 5, 8, 13, 21, 34, 55, 89, 144, etc.
Fn = Fn-1+ Fn-2
F0 = 0, F1 = 1
sequence5.pl
print "Please type a DNA sequence: ";
my $DNAseq = <>;
chomp($DNAseq);
#Copy the sequence to a new variable
my $asDNAseq = $DNAseq;
#'translate' a->t, c->g, g->c, t->a
$asDNAseq =~ tr/acgt/tgca/;
print "Complementary strand:\n$asDNAseq\n";
$asDNAseq =~ tr/acgt/tgca/;
=~ is a binding operator and means: perform the following
action on this variable.
The operation tr/// translates each character from the first set
of characters into the corresponding character in the second set:
acgt
||||
tgca
Counting
tr/// can also be used to count characters. If the
second part is left empty, no translation takes place.
$numberOfNs = ($DNASeq =~ tr/N//);
'automatic' typing
using a pipe "|":
echo ggatcc | perl sequence5.pl
or redirect using "<":
perl sequence5.pl < sequence.txt
Exercise 1.
Create a program that reads a DNA sequence
from the keyboard, and reports the sequence
length and the G/C content of the sequence (as
a fraction)
perltidy
program that properly formats your perl script
Indentation, spaces, etc.
perltidy yourscript.pl
Result is in:
yourscript.pl.tdy
0
1
2
@months
3
a list variable or array starts with
an @ followed by its name
Arrays
my @fibonacci = (0,1,1,2);
print @fibonacci;
print $fibonacci[3];
$fibonacci[4] = 3;
$fibonacci[5] = 5;
$fibonacci[6] = 8;
@fibonacci
0
0
1
1
2
1
3
2
Arrays
my @hw = ("Hello ","World","\n");
print @hw;
my @months = ( "January",
"February",
"March");
Arrays
To access a single element of the list use the
array name with $ instead of the @ and append
the position of the element in: [ ]
print $months[1];
February
$hw[1] = "Wur";
print @hw;
Arrays
To find the index of the last element in the list:
print $#months;
2
To find the number of elements in an array:
print $#months + 1;
or:
print scalar(@months);
Arrays
Note: like many programming languages, the
index of the first item in an array is not 1, but 0!
Note: $months ≠ $months[0] !!!
Growing and shrinking arrays
push:
pop:
shift:
unshift:
splice:
add an item to the end of the list
remove an item from the end of the list
remove an item from the start of the list
add an item to the start of the list
insert/remove one or more items
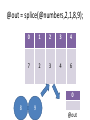
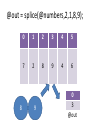
@out = splice(@array, start, length, @in);
@numbers
index
0
1
2
3
4
value
1
2
3
4
5
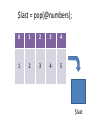
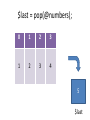
$last = pop(@numbers);
0
1
2
3
4
1
2
3
4
5
$last
$last = pop(@numbers);
0
1
2
3
1
2
3
4
5
$last
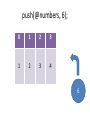
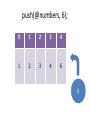
push(@numbers, 6);
0
1
2
3
1
2
3
4
6
push(@numbers, 6);
0
1
2
3
4
1
2
3
4
6
6
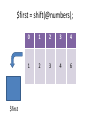
$first = shift(@numbers);
$first
0
1
2
3
4
1
2
3
4
6
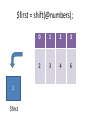
$first = shift(@numbers);
1
$first
0
1
2
3
2
3
4
6
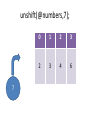
unshift(@numbers,7);
7
0
1
2
3
2
3
4
6
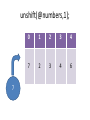
unshift(@numbers,1);
7
0
1
2
3
4
7
2
3
4
6
@out = splice(@numbers,2,1,8,9);
0
1
2
3
4
7
2
3
4
6
0
8
9
@out
@out = splice(@numbers,2,1,8,9);
8
0
1
2
3
4
5
7
2
8
9
4
6
9
0
3
@out
my ($x,$y,$z) = @coordinates;
my @words = split(" ", "Hello World");
$words[0] = "Hello"
$words[1] = "World"
More loops
my @plantList = ("rice", "potato", "tomato");
print $plantList[0];
print $plantList[1];
Print $plantList[2];
Or:
foreach my $plant (@plantList) {
print $plant;
}
Loops
foreach variable ( list ) {
do something with the variable
}
foreach my $i ( @lotto_numbers ) {
print $i;
}
foreach my $i ( 1 .. 10, 20, 30 ) {
print $i;
}
Loops
for variable ( list ) {
do something with the variable
}
for my $i ( 1, 2, 3, 4, 5 ) {
print $i;
}
for my $i ( 1 .. 10, 20, 30 ) {
print $i;
}
Loops
while ( condition ) {
do something
}
my $i = 0;
while ($i < 10) {
print "$i < 10\n";
$i++;
}
Loops
for ( init; condition; increment ) {
do something
}
for (my $i = 0; $i < 10; $i++) {
print "$i < 10\n";
}
Loops
my $i = 0;
while ($i < 10) {
print "$i < 10\n";
$i++;
}
for (my $i = 0; $i < 10; $i++) {
print "$i < 10\n";
}
Exercise
Write a script that reverses a DNA sequence
use an array
Hint: Splitting on an empty string "" splits after
every character.
@sequence = split("",$sequence);
0
1
Name
Box
Crick
3
Franklin
1
Watson
0
Wilkins
2
2
%phonebook
3
a hash table variable starts with a %
followed by its name
Hash tables
Also called lookup tables, dictionaries or
associative arrays
key/value combinations: keys are text, values
can be anything
%month_days = ("January" => 31,
"February" => 28,
"March" => 31 );
Hash tables
To access a value in the hash table, use the hash
table name with $ instead of the % and append
the key between { }
$month_days{"February"} = 29;
print $month_days{"January"};
31
Hash tables
The 'keys' function returns an list with the keys of
the hash table. There is also a 'values' function.
@month_list = keys(%month_days);
# ("January", "February", "March")
Hash tables
my %latin_name=(
"rice" => "Oryza sativa",
"potato" => "Solanum tuberosum"
)
foreach my $common_name (keys(%latin_name)){
print "$common_name: " ;
print "$latin_name{$common_name}\n";
}
rice: Oryza sativa
potato: Solanum tuberosum
Hash tables
The keys have to be unique, the values do not.
The order of elements in a hash table is not
reliable, first in is not necessarily first out.
You can use 'sort' to get the keys in an
alphabetically ordered list:
@sorted = sort(keys(%latin_name));
Exercise
Create a hash table with codons as keys and the
corresponding amino acids as the values
Hint: search for the standard genetic code in the
"genetic code" database at:
http://srs.bioinformatics.nl/
Use the three lines for the first, second and third
base and the line for the corresponding AA.
I/O: Input and Output
reading and writing files
Reading and writing files
open FASTA, "sequence.fa";
my $firstLine = <FASTA>;
my $secondLine = <FASTA>;
close FASTA;
Reading and writing files
Files need to be opened before use
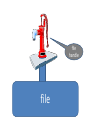
Reading and writing files
Perl uses so-called “file handles” to attach to
files for reading and writing
file
handle
file
Opening files
General
open FileHandle, "mode", "filename"
Open for reading:
open LOG, "<", "/var/log/messages";
open LOG, "/var/log/messages";
Open for writing:
open WRT, ">", "newfile.txt";
Open for appending:
open APP, ">>", "existingfile.txt";
Defensive programming
my $fastaName = "sequence.fa";
open FASTA, $fastaName or
die "cannot open $fastaName\n";
Reading from a file
reading from an open file via the filehandle:
$firstLine = <FASTA>;
$secondLine = <FASTA>;
@otherLines = <FASTA>;
<FASTA>
Reads one line if the result goes into a scalar
$line = <FASTA>;
Reads all (remaining) lines if the result goes into
an array
@lines = <FASTA>;
file handles 'remember' the position in the file
Standard in and standard out
The keyboard and screen also have 'file' handles,
remember STDIN and STDOUT
read from the keyboard:
$DNAseq = <STDIN>;
write to the screen:
print STDOUT "Hello World\n";
Reading a file
open FASTA, "sequence.fa" or die;
my $sequence = "";
while (my $line = <FASTA>) {
chomp($line);
$sequence .= $line;
}
close FASTA;
print $sequence,"\n";
(my $line = <FASTA>)
also is a condition
true: line could be read
false: EOF, end of file
Identical?
while (my $line =
print $line;
}
<FASTA>) {
for my $line (<FASTA>) {
print $line;
}
Not completely
Read line by line:
while (my $line = <FASTA>) {
print $line;
}
First read complete file into computer memory:
for my $line (<FASTA>) {
print $line;
}
Writing to a file
open RANDOM, ">", "Random.txt";
for(1..50) {
my $random = rand(6);
print RANDOM "$random\n";
}
close RANDOM;
Writing to a file
open RANDOM, ">", "Random.txt";
for(1..50) {
my $rnd = rand(6);
$rnd = sprintf("%d\n",$rnd + 1);
print RANDOM $rnd;
}
close RANDOM;
Closing the file
close filehandle;
close FASTA;
A file is automatically closed if you (re)open a
file using the same filehandle, or if the Perl
script is finished.
Minimalistic Perl
open FASTA, "sequence.fa" or die;
my $sequence = "";
while (my $line = <FASTA>) {
chomp($line);
$sequence .= $line;
}
close FASTA;
print $sequence,"\n";
Minimalistic Perl
open FASTA, "sequence.fa" or die;
my $sequence = "";
while (<FASTA>) {
chomp;
$sequence .= $_;
}
close FASTA;
print $sequence,"\n";
$_
default scalar variable, if no other
variable is given. But only in selected
cases...
Minimalistic Perl
open FASTA, "sequence.fa" or die;
my $sequence = "";
while (<FASTA>) {
chomp;
$sequence .= $_;
}
close FASTA;
print $sequence,"\n";
Minimalistic Perl
open FASTA, "sequence.fa" or die;
my $sequence = "";
while ($_ = <FASTA>) {
chomp($_);
$sequence .= $_;
}
close FASTA;
print $sequence,"\n";
Exercises
2. Adapt the G/C script so multiple sequences in
FASTA format are read from a file
3. Modify the script to process a file containing
any number of sequences in EMBL format
4. Now let the program generate the reverse
complement of the sequence(s), and report
sequence length and G/C content
Exercises
5. Use the rand function of Perl to shuffle the
nucleotides of the input sequence, while
maintaining sequence composition; again
report sequence length and G/C content