* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download CheW
Survey
Document related concepts
Transcript
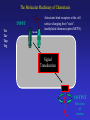
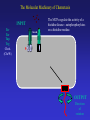
Sept 25 Biochemical Networks Chemotaxis and Motility in E. coli Examples of Biochemical and Genetic Networks • Background • Chemotaxis- signal transduction network Bacterial Chemotaxis Flagellated bacteria “swim” using a reversible rotary motor linked by a flexible coupling (the hook) to a thin helical propeller (the flagellar filament). The motor derives its energy from protons driven into the cell by chemical gradients. The direction of the motor rotation depends in part on signals generated by sensory systems, of which the best studied analyzes chemical stimuli. Chemotaxis - is the directed movement of cells towards an “attractant” or away from a “repellent”. • For a series of QuickTime movies showing swimming bacteria with fluorescently stained flagella see: http://www.rowland.org/bacteria/movies.html • For a review of bacterial motility see Berg, H.C. "Motile behavior of bacteria". Physics Today, 53(1), 24-29 (2000). (http://www.aip.org/pt/jan00/berg.htm) A photomicrograph of three cells showing the flagella filaments. Each filament forms an extend helix several cell lengths long. The filament is attached to the cell surface through a flexible ‘universal joint’ called the hook. Each filament is rotated by a reversible rotary motor, the direction of the motor is regulated in response to changing environmental conditions. The E. coli Flagellar Motor- a true rotary motor Rotationally averaged reconstruction of electron micrographs of purified hook-basal bodies. The rings seen in the image and labeled in the schematic diagram (right) are the L ring, P ring, MS ring, and C ring. (Digital print courtesy of David DeRosier, Brandeis University.) Tumble (CW) Smooth Swimming or Run (CCW) Chemotactic Behavior of Free Swimming Bacteria No Gradient Increasing attractant Increasing repellent A ‘Soft Agar’ Chemotaxis Plate A mixture of growth media and a low concentration of agar are mixed in a Petri plate. The agar concentration is not high enough to solidify the media but sufficient to prevent mixing by convection. The agar forms a mesh like network making water filled channels that the bacteria can swim through. A ‘Soft Agar’ Chemotaxis Plate Bacteria are added to the center of the plate and allowed to grow. A ‘Soft Agar’ Chemotaxis Plate As the bacteria grow to higher densities, they generate a gradient of attractant as they consume it in the media. Attractant Concentration cells cells A ‘Soft Agar’ Chemotaxis Plate The bacteria swim up the gradients of attractants to form ‘chemotactic rings’ . This is a ‘macroscopic’ behavior. The chemotactic ring is the result of the ‘averaged” behavior of a population of cells. Each cell within the population behaves independently and they exhibit significant cell to cell variability (individuality). A ‘Soft Agar’ Chemotaxis Plate ‘Serine’ ring ‘Aspartate’ ring Each ‘ring’ consists of tens of millions of cells. The cells outside the rings are still chemotactic but are just not ‘experiencing’ a chemical gradient. Serine and aspartate are equally effective “attractants”, but in this assay the attractant gradient is generated by growth of the bacteria and serine is preferentially consumed before aspartate. Assays of Bacterial Motility Brownian Motion Latex Beads Swimming E. coli Fluorescent Flagella Bundle Tethered E. coli Tracking E. coli Assays of Bacterial Motility Flow Chamber Assay Surface Swarming Salmonella Pattern Formation Laser Trap The Molecular Machinery of Chemotaxis INPUT Attractant concentration Signal Transduction OUTPUT Direction of rotation The Molecular Machinery of Chemotaxis INPUT Tsr Tar Tap Trg Attractants bind receptors at the cell surface changing their “state”. (methylated chemoreceptors MCPS). Signal Transduction OUTPUT Direction of rotation The Molecular Machinery of Chemotaxis INPUT Tsr Tar Tap Trg CheA (CheW) The MCPs regulate the activity of a histidine kinase - autophosphorylates on a histidine residue. P~ OUTPUT Direction of rotation The Molecular Machinery of Chemotaxis CheA transfers its phosphate to a signaling protein CheY to form CheY~P. INPUT Tsr Tar Tap Trg CheA (CheW) CheY P~ P~ OUTPUT Direction of rotation The Molecular Machinery of Chemotaxis CheY~P binds to the “switch” and causes the motor to reverse direction. The signal is turned off by CheZ which dephosphorylates CheY. INPUT Tsr Tar Tap Trg CheA (CheW) CheY CheZ P~ P~ OUTPUT Direction of rotation Excitatory Pathway At ‘steady state’, CheY~P levels in the cell are constant and there is some probability of the cell tumbling. Binding of attractant of the receptorkinase complex, results in decreased CheY~P levels and reduces the probability of tumbling and the bacteria will tend to continue in the same direction. MCP CheA (CheW) CheY~P CheZ Motor + attractant CheY inactive The Molecular Machinery of Chemotaxis Adaptation involves two proteins, CheR and CheB, that modify the receptor to counteract the effects of the attractant. INPUT Tsr Tar Tap Trg CheA (CheW) CheY CheZ CheR CheB P~ P~ OUTPUT Direction of rotation Adaptation Pathway CheR MCP CheA (CheW) Less active CheB~P MCP~CH3 CheA (CheW) More active Adaptation Pathway CheR MCP-(CH3)0 MCP-(CH3)1 MCP-(CH3)2 MCP-(CH3)3 MCP-(CH3)4 MCP-(CH3)0 MCP-(CH3)1 MCP-(CH3)2 MCP-(CH3)3 MCP-(CH3)4 +Attractant +Attractant +Attractant +Attractant +Attractant CheB~P In a receptor dimer there will 65 possible states (5 methylation states and two occupancy states per monomer). If receptors function in receptor clusters, essentially a continuum of states may exist. Some Issues in Chemotaxis: • Sensing of Change in Concentration not absolute concentration i.e. temporal sensing • Exact Adaptation • Sensitivity and Amplification • Signal Integration from different Attractants/Repellents The range of concentration of attractants that will cause a chemotactic response is about 5 orders of magnitude (nM mM) References on Modeling Chemotaxis Barkai, N. & Leibler, S. (1997) Nature (London) 387: 913–917. Spiro, P. A., Parkinson, J. S. & Othmer, H. G. (1997) Proc. Natl. Acad. Sci. US 94: 7263–7268. Tau-Mu Yi, Yun Huang , Melvin I. Simon, and John Doyle (2000) Proc. Natl. Acad. Sci. USA 97: 4649–4653.* Bray, D., Levin, M. D. & Morton-Firth, C. J. (1998) Nature (London) 393: 85–88. * * - these models have incorporated the Barkai model. Robustness in simple biochemical networks N. Barkai & S. Leibler Departments of Physics and Molecular Biology, Princeton University, Princeton, New Jersey 08544, USA Simplified model of the chemotaxis system. Mechanism for robust adaptation E is transformed to a modified form, Em, by the enzyme R; enzyme B catalyses the reverse modification reaction. Em is active with a probability of am(l), which depends on the input level l. Robust adaptation is achieved when R works at saturation and B acts only on the active form of Em. Note that the rate of reverse modification is determined by the system’s output and does not depend directly on the concentration of Em (vertical bar at the end of the arrow). Some parameters used to characterize the network. Tumble frequency Steady-State Tumble Frequency Adaptation Time Adaptation precision Chemotactic response and adaptation in the Model. The system activity, A, of a model system which was subject to a series of step-like changes in the attractant concentration, is plotted as a function of time. Attractant was repeatedly added to the system and removed after 20 min, with successive concentration steps of l of 1, 3, 5 and 7 mM. Note the asymmetry to addition compared with removal of ligand, both in the response magnitude and the adaptation time. How robust is the model with respect to variation in parameters? Adaptation precision Adaptation Time Adaptation precision (i.e. exact adaptation) is Robust Adaptation time is very sensitive to parameters Testing the predictions of the Barkai model Robustness in bacterial chemotaxis. U. Alon, M. G. Surette, N. Barkai & S. Leibler • The concentration of che proteins were altered as a simple method to vary network parameters. • The behavior of the cells were measured (adaptation precision, adaptation time and steady-state tumble frequency). • In each case the predictions of the model we observed. Data for CheR As predicted by the model the adaptation precision was robust while adaptation time and steady-state tumble frequency were very sensitive to conditions.