* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download The Major Histocompatibility Complex
Designer baby wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Nicotinic acid adenine dinucleotide phosphate wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
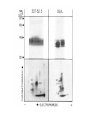
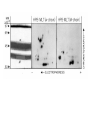
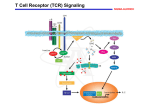
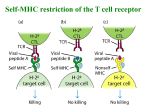
TCR Identification February 13, 2007 The elusive TCR “The T-cell receptor-the molecule in the T-cell membrane capable of specifically binding antigen-is the San Andreas Fault of immunology: here the plates rub against each other, shifting gradually and unnoticed, and a major “earthquake” may occur at any moment.” Klein 1982 “Few issues in science have proved as difficult to resolve and as subject to contention as the nature of the TCR.” Jean Marx (writer for Science) 1983 “The study of the T cell receptor has been the subject of many studies, many of which have led to diverse and controversial findings. At the present time, the consensus is that the receptor is not conventional surface Ig as found on B cells.” Unanue and Benacerraf 1984 Initial lack of success in identifying the TCR • IgT-antibodies against an Ig-like molecule on the surface of T cells • Identify/clone by homology to immunoglobulin genes IgT Identifying the TCR • Two overall approaches – cDNA cloning – anti-TCR antibodies, protein • Leading laboratories – cDNA:Hedrick/Davis (mouse), Tak Mak (human) – Protein: Kappler/Marrack,Allison, Trowbridge, Reinherz Isolation of cDNA clones encoding T cell-specific membraneassociated proteins Steve Hedrick, David Cohen, Ellen Nielsen, and Mark Davis Laboratory of Immunology NIAID, NIH Cytochrome c • Model protein with isolates from many different species to study the fine specificity of T cell recognition • Ron Schwartz’s laboratory at NIH was a leading investigator studying cytochrome c • Canonical response was B10.A mice against pigeon cytochrome c (PCC) • A dominant epitope 91-104 of PCC was identified that was I-Ek restricted • A heteroclitic response of the same T cells was observed with moth cytochrome c 91-103 • The heteroclitic response was due to the MCC peptide being one AA shorter than PCC, which resulted in an optimal I-Ek anchor residue at P9 • The response to MCC is oligoclonal and utilizes V3 and V11 exclusively Sequence of TM86 clone reveals an immunoglobulin like molecule LOCUS 1109237A 123 aa DEFINITION T cell receptor VDJbeta 1.9.2. ACCESSION 1109237A VERSION 1109237A GI:224610 DBSOURCE prf: locus 1109237A; linear VRL 26-JUN-1996 state: germ; taxonomy: Virus. KEYWORDS T Cell Receptor V beta Gene; Mouse; Seq Determination; 450bp; 96AAs; Use Limited Repertoire of Gene; Somatic Diversification at V-D-J Region; DNA Blot; Single Gene Subfamily; Variability Plot; Hydrophobicity; beta Sheet Potential. SOURCE Murid herpesvirus 1 (Murine cytomegalovirus) ORGANISM Murid herpesvirus 1 Viruses; dsDNA viruses, no RNA stage; Herpesviridae; Betaherpesvirinae; Muromegalovirus. REFERENCE 1 (residues 1 to 123) AUTHORS Barth,R.K., Kim,B.S., Lan,N.C., Hunkapiller,T., Sobieck,N., Winoto,A., Gershenfeld,H., Okada,C., Hansburg,D., Weissman,I.L. and Hood,L. TITLE The murine T-cell receptor uses a limited repertoire of expressed V beta gene segments JOURNAL Nature 316 (6028), 517-523 (1985) PUBMED 2412120 COMMENT cDNA. FEATURES Location/Qualifiers source 1..123 /organism="Murid herpesvirus 1" /db_xref="taxon:10366" Region 30..123 /region_name="IGv" /note="Immunoglobulin domain variable region (v) subfamily; members of the IGv subfamily are components of immunoglobulins and T-cell receptors; cd00099" /db_xref="CDD:28982" ORIGIN 1 vslclvetal mntkitqspr ylilgranks leceqhlghn amywykqsae kppelmflyn 61 lkqlirnetv psrfipecpd ssklllhisa vdpedsavyf cassqtggnt evffgkgtrl 121 tvv Blast search of TM86 identifies it to be V1, but cytochrome c responses are all V3! Cloning of the human TCR Complete primary structure of a heterodimeric T-cell receptor deduced from cDNA sequences. * Saito H, * Kranz DM, * Takagaki Y, * Hayday AC, * Eisen HN, * Tonegawa S. Nature. 1984 Jun 28-Jul 4;309(5971):757-62. Two related, but distinct, cDNA clones have been isolated and sequenced from a functional murine cytotoxic T-lymphocyte clone. The genes corresponding to these cDNA are expressed and rearranged specifically in T cells and both have similarities to immunoglobulin variable and constant region genes. It is concluded that these genes code for the two subunits of the heterodimeric antigen receptor on the surface of the T cell; its complete deduced primary structure is presented. PMID: 6330561 [PubMed - indexed for MEDLINE] Transfection of these two receptors does not recapitulate the specificity of the 2C TCR! The Major Histocompatibility Complex-restricted on T cells in Mouse and Man: Identification of Constant and Variable Peptides John Kappler, Ralph Kubo, Kathryn Haskins, Charles Hannum, Michele Pigeon, and Pippa Marrack National Jewish Hospital, Denver Brad McIntyre and Jim Allison UT, Smithville-> Berkeley->MSK Ian Trowbridge Salk Institute, La Jolla Major players in the protein identification of the TCR • Kappler and Marrack – Generated monoclonal antibodies against the TCR of ovalbumin specific T cell hybridomas (i.e. DO.11.10) • Jim Allison – Generated a monoclonal antibody against a T cell tumor line • Ian Trowbridge – Found an anti-TCR in a mouse anti-human T cell monoclonal antibody-retrospectively • Ellis Reinherz – Monoclonal against an alloreactive human clone A T cell name REX “REX line was obtained from a patient with T cell lymphoblastic lymphoma which gave rise to spontaneously growing subclones as isolated by limiting dilution technique.”