* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Survey
Document related concepts
Transcript
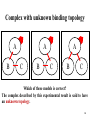
Advanced Bioinformatics Lecture 3: Protein-protein interaction ZHU FENG [email protected] http://idrb.cqu.edu.cn/ Innovative Drug Research Centre in CQU 创新药物研究与生物信息学实验室 Table of Content 1. Protein-protein interaction 2. Interaction representations 3. Method A: Two-hybrid assay 4. Method B: Affinity purification 5. Spoke and matrix models of PPI 2 Protein–protein interaction (PPI) The horseshoe shaped ribonuclease inhibitor (shown as wireframe) forms a protein– protein interaction with the ribonuclease protein. The contacts (non-covalent interaction) between the two proteins are shown as colored patches. 3 Central importance for processes in cell Signal transduction: signals from the exterior of a cell are mediated inside by PPI of the signaling molecules. Protein transportation: from cytoplasm to nucleus or vice versa in the case of the nuclear pore importins. Protein modification: a protein kinase will add a phosphate to a target protein. Chain interaction: proteins with SH2 domains only bind to other proteins when they are phosphorylated on the amino acid tyrosine while specifically recognize acetylated lysines. 4 Interaction representation Enzyme + Substrate Kinase-ATP complex + inactive-enzyme ==> Kinase + ADP + active enzyme K P ATP ADP One representation 5 Interaction representation Inactive enzyme Kinase-ATP complex Active enzyme ADP Another representation 6 Generalization of the representation A C B D E … F A biomolecule’s function can be defined by the things that it interacts Makes it easy focus on the interaction part with and the new (or toaltered) molecules that result from that interaction. 7 A simple record A 01. Short label for A 03. Molecule type for A 05. Database reference for A 07. Where A comes from 09. Interaction Kinetics 10. Publication reference B 02. Short label for B 04. Molecule type for B 06. Database reference for B 08. Where B comes from The minimal record has 10 pieces of information 8 An example record A B 01. EGF 02. EGFR 03. Protein 04. Protein 05. OMIM: 131530 06. OMIM: 131550 07. Homo sapiens 08. Homo sapiens 09. Equilibrium dissociation constant (Kd) = 130 nM 10. Cancer Cell 7(4):301-311, 2005 You can view this record in BIND (http://bind.ca/) with ID: 263509 9 BIND stores molecular interaction data 10 BIND interaction types Specify method used to confirm the interaction, what method? 11 Methods for detecting interactions Many interactions in BIND originates from high-throughput experiments designed to detect interactions between proteins The most common methods are – Two-hybrid assay – Affinity purification 12 Methods comparison Yeast two hybrid screens allow for interactions between proteins that are never expressed in the same time and place, lowering the specificity, but better indicate non-specific tendencies towards sticky interactions Affinity purification better indicates functional in vivo protein-protein interactions. 13 Method A: Two-hybrid assay Transcription activation domain (AD) 1. Transcriptional activator (TA) 2. 3. Gene Promoter DNA-binding domain (BD) 4. Fields S, et al. Nature. 1989 Jul 20;340(6230):245-6. 14 Two-hybrid assay SNF4 1. SNF1 UASG B A 2. 3. Reporter Gene 4. 15 Two-hybrid assay 16 Two-hybrid assay 17 Some two-hybrid caveats 1. A 2. 3. 4. Does the DBD-fusion have activity by itself? 18 Some two-hybrid caveats 1. C B A 2. 3. 4. Is the ‘interaction’ mediated by some other protein? 19 Some two-hybrid caveats 1. B A 2. 3. 4. Is the ‘interaction’ bi-directional? 20 Method B: Affinity purification A Protein of interest This molecule will bind the ‘tag’ Tag modification (e.g. HA/GST/His) 21 Affinity purification The cell A 22 Affinity purification Lots of other untagged proteins A B Naturally binding protein 23 Affinity purification Ruptured membranes A B Cell extract 24 Affinity purification A B Untagged proteins go through fastest (flow-through) 25 Affinity purification A B Tagged complexes are slower and come out later (eluate) 26 Some affinity purification caveats Firstly, this is only a representation of observation A You can only tell what proteins are in the eluate B You can’t tell how they are connected If there is only one other protein present (B), then its likely that A and B are directly interacting A B C But, what if I told you that two other proteins (B and C) were present along with A … 27 Complex with unknown binding topology A B A C B A C B C Which of these models is correct? The complex described by this experimental result is said to have an unknown topology. 28 Complexes with unknown stoichiometry A B A C Here’s another possibility? The complex described by this experimental result is also said to have unknown stoichiometry. 29 Spoke and matrix models of PPI Spoke Simple, intuitive, more accurate, but can misrepresent Possible Actual Topology Matrix Theoretical max. no. of interactions, but many FPs Bader GD, et al. Nat Biotechnol. 2002 20(10):991-7. 30 Synthetic genetic interactions in yeast Cell Polarity Cell Wall Maintenance Cell Structure Mitosis Chromosome Structure DNA Synthesis DNA Repair Unknown Others 31 Network of the human interactome Each point represents a protein and each line between them is an interaction 32 Network motifs found in networks The feed-forward loop, bi-fan and biparallel are over-represented, whereas feedback loop is under-represented in gene regulatory networks and neuronal connectivity networks. 33 Yeast interactome project 34 Interactome data analysis (1) Validation of interactome’s coverage and quality Interactomes are never complete, given the limitations of experimental methods. For instance, it has been estimated that typical Y2H screens detect only 25% or so of all interactions in an interactome. The coverage of an interactome can be assessed by comparing it to benchmarks of well-known interactions that have been found and validated by independent assays. 35 Interactome data analysis (2) Protein function prediction Assumption: uncharacterized proteins have similar functions as their interacting proteins. For example, YbeB with unknown function was found to interact with ribosomal proteins and later shown to be involved in translation. Although such predictions may be based on single interactions, usually several interactions are found. Thus, the whole network of interactions can be used to predict protein functions, given that certain functions are usually enriched among the interactors. 36 Interactome data analysis (3) Perturbations and disease The topology of an interactome makes it possible to predict how a network reacts to the perturbation (e.g. removal) of nodes (proteins) or edges (interactions). Mutations of genes (and thus their proteins) can cause perturbations of networks and thus disease. 37 Interactome data analysis (4) Network structure and modules The distribution of properties among the proteins of an interactome has revealed functional modules within a network that indicate specialized subnetworks. Such modules can be purely functional, as in a signaling pathway, or structural, as in a protein complex. In fact, it is a formidable task to identify protein complexes in an interactome, given that typically no affinities are known. 38 Projects Q&A! 1. Biological pathway simulation 2. Computer-aided anti-cancer drug design 3. Disease-causing mutation on drug target Any questions? Thank you! 39