* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Multiple perturbation analysis of cancer pathways
Survey
Document related concepts
Transcript
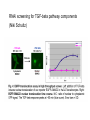
How about a Master’s project in the area of cancer systems biology? • We offer projects in the following areas: • Reverse engineering of cancer pathways • Control of the cancer cell phenotype using drug combinations • Predicting phenotype responses to cellular perturbation from array data • Contact for more info: • [email protected] (please attach a CV) • Previous project workers: • • • • Frank Eriksson, Mat Stat, Chalmers Darima Lamazhapova, Cambridge University Erik Larsson, Wallenberg lab, GU Tanya Lobovkina, Chemistry, Chalmers Multiple Perturbation Analysis of Cancer Pathways Sven Nelander Computational Biology Center / Chris Sander group Memorial Sloan-Kettering Cancer Center New York Outline • Perturbing cancer cells - key questions • Building models for combinatorial perturbation of breast cancer pathways • Whole-genome RNAi screening to extend the TGF-beta pathway • Work in cancer genomics and related areas Cancer • A broad class of diseases exhibiting uncontrolled growth, tissue invasion and metastasis. • Gradual progression towards a more malignant phenotype • Acquisition of mutations that affect a specific set of processes • • • • • Growth factor signaling Apoptosis DNA repair Cell cycle regulation Differentiation Selective targeting of cancer pathways by small compounds 1. Force differentiation 2. Inhibit anti-apoptotic signals 3. Inhibit growth-stimulating signals Tumors contain multiple genetic abnormalities • Copy number alterations (MSKCC study). Gain of DNA 200 Patients Loss of DNA genomic position (3000 megabases) • Sequence alterations. 90 mutated genes per tumor in breast and colon cancer (Sjöblom et al, Science 2006) • Promoter hypermethylation. A role for systems biology? • Key types of question: • • • • How will a melanoma cell line with mutation X respond to drug Y? Will drug X synergize with drug Y? Which regulatory interactions are implicated by the observed responses? What’s the mechanism of action of drug X? • Pathway maps are ambiguous, incomplete and have unclear predictive value. • Expert intuition is likely to fail in complicated cases. • To facilitate prediction and inference, mathematical models can be employed. Implementing a systems biology cycle for combined perturbations Desirable properties of a model for our purposes Representational capability: • Pathway-like and biochemically plausible • Quantitative or semi-quantitative predictions • Nonlinear interaction effects (epistasis and synergism) are possible Experimental implications: • Both temporal and steady state perturbation responses • Incomplete readout possible Algorithms: • Reverse engineering is computationally tractable Simple dynamical models dx i (W ij x j ) i x i Pi dt j Similar models used for Analysis of microarray time series (D’Haeseler 2000, Xiong 2004) Network inference from perturbed microarray profiles (Yeung 2002, Tegner 2003) Inference of mechanism of action (diBernardo et al 2006) dx i i f (W ij x j Pi ) i x i dt j Similar models used for Analysis of microarray time series (D’Haeseler, 2000), Modeling of lambda phage gene regulation (Vohradsky, 2001), Robustness analysis of the yeast cell cycle (Li et al 2004). Discussed as a model for signaling in (Bhalla 2003). DNA switch network - synthetic biology (Kim et al 2004 and 2006) Prediction of perturbation responses COMPOUNDS PHENOTYPE Prediction of perturbation responses Experimental data from Kaufman et al, PLoS Comp Biol, 2006 Parameter fitting / system identification • For all experiments minimize E E SSQ E STRUCT Sum of squares error Solmaz Shahalizadeh, Master’s thesis Structural complexity Algorithms used to minimize E • Recurrent backpropagation (Pineda, 1988) • Backpropagation through time (Pearlmutter) • Gennemark and Wedelin, 2007 Inference from steady state perturbation responses, hypothetical experiment with 40 dual perturbations and 10 readouts Inference from perturbation responses, experimental data Data from Janes et al, Science 2005 Experimental pilot studies (ongoing) • Two breast epithelial cell lines • MCF7 - cancer • MCF10A - transformed noncancer • Initial focus on mitogenic pathways and low molecular weight compound perturbation • Database of 2200 compound-gene links • Experiment 1: predict triplet perturbations from dual perturbations • Experiment 2: crosstalk detection and explanation Efficient proteomics technique will make large perturbation studies possible. SILAC technology (Jens Andersen group, Odense) Reverse phase protein array (Weiqing Wang) Dilution of Lysate 0 1/2 1/4 1/8 Duplicates 0 1/2 1/4 1/8 1/16 1/32 1/64 1/128 1/16 1/32 1/64 1/128 Duplicates 1) One grid for one sample 2) One antibody blot for one slide 3) Relative quantification, positive controls on each slide 4) Quantitative peptide and phosphopeptide controls RNAi screening for TGF-beta pathway components (Niki Schultz) 21000 siRNA duplexes were scored for their effect on TGFbeta signaling Work in genomics • Sarcoma genome project • Collaboration with MSKCC surgery dept and Broad Inst. • 140 sarcoma patients • Large-scale genomic characterization: – Transcriptional arrays – Copy number arrays – Exon sequencing • Aberrant processes? Therapy targets? • DNA copy number alteration in nonmalignant lesions • Collaboration with Columbia pathology dept. Summary • Methodology to analyze combinatorial perturbation experiments using differential equation models. • Preliminary data suggest applicability to real experimental data • No assumptions of linearity or complete observation • The methodology generalizes genetic epistasis analysis in that it handles higher order perturbations and feedback loops. • We are proceeding to a study of combinatorial drug effects on the phenotype of breast cancer cells. Future perspectives • Using perturbation to pinpoint mutations and regulatory differences between tumors • Cancer genomics data as an endogenous perturbation experiment • Phenotype control in non-malignant disease conditions Acknowledgements • Weiqing Wang, Nikolaus Schultz, Christine Pratilas, Barry Taylor, Dina Marenstein, Sam Singer, Joan Massague, Neal Rosen, Chris Sander • Solmaz Shahalizadeh, Peter Gennemark, Frank Eriksson, Darima Lamazhapova • Søren Schandorff, Jens Andersen • Björn Nilsson