* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Immune Functions of B Cells
Drug discovery wikipedia , lookup
Drug design wikipedia , lookup
Neuropsychopharmacology wikipedia , lookup
Drug interaction wikipedia , lookup
Prescription costs wikipedia , lookup
NK1 receptor antagonist wikipedia , lookup
Neuropharmacology wikipedia , lookup
Pharmacognosy wikipedia , lookup
Pharmacokinetics wikipedia , lookup
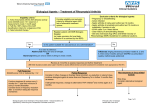
Expanding the Research Domains of Rheumatoid Arthritis Clinical Databases: The Promise of Pharmacogenetics Jeffrey Greenberg, MD, MPH NYU Hospital for Joint Diseases New York University School of Medicine December 2006 Outline • Why Pharmacogenetics • Case Study: TNF Antagonists • Issues of Study Design and Analysis • Future Directions How Variable is the Human Genome? Size: 3 billion base pairs of DNA Content: 39,114 genes Variation: 1% of base pairs are polymorphic (i.e. 30 million base pairs) Pharmacogenetics and “Personalized” Medicine Marsh, S. and McLeod HL. Hum. Mol. Genet. 2006 15:R89-93. Why Is Pharmacogenetics Important? “ Let us […] imagine the despair of the mouse experimentalist when we suggest that he or she randomly allocate treatments to animals of different genetic backgrounds and immunized with different disease triggers.” ….[This is essentially how we treat RA in 2006] Klareskog, L. Nature Clinical Practice Rheumatology (2006) 2, 517. Arthritis Clinical Databases and Pharmacogenetics • 6 biologic agents have been FDA approved for RA. • Only 1 RCT (Early RA Trial) has published pharmacogenetic studies. • Arthritis clinical databases may represent the ONLY practical approach to advancing the field of pharmacogenetics. • As more biologic agents are approved, biomarkers that can predict response will be increasingly important. GENETIC POLYMORPHISMS Pharmacokinetic •Transporters •Plasma protein binding •Metabolism Pharmacodynamic •Receptors •Ion channels •Enzymes •Immune molecules Potential Consequences of Different Drug Metabolism and Drug Receptor Genotypes Evans WE and Relling MV. Science 286:487-491, 1999. Polymorphic Drug Metabolizing Enzymes ALDH2 Acetaldehyde dehydrogenase CYP1A2 Caffeine, acetaminophen demethylation CYP2C9 Warfarin hydroxylation CYP2C19 Mephenytoin hydroxylation CYP2D6 Debrisoquine hydroxylation UGT2 Ibuprofen and naproxen glucuronidation Isoniazide acetylation NAT 2 Proof of Principle: Warfarin and Cytochrome P450 Cyp2C9 Alleles Dervieux T et al, Mutat Res 2005:180-194. Methylene Tetrahydrofolate Reductase (MTHFR) Dervieux T et al, Mutat Res 2005:180-194. Effect of the MTHFR C677T Polymorphism on Methotrexate Toxicity in RA Patients Urano W et al. Pharmacogenetics 2002: 12(3): 183-190 Pharmacogenetics of TNF Antagonists for the Treatment of RA Case Study How Can We Predict Response (or Failure) of Methotrexate or TNF Antagonists in RA Patients? ACR20 Response ACR50 Response 90 80 ACR70 Response 85 76 75 69 70 60 50 48 43 43 40 30 24 19 20 10 0 MTX Etanercept Etanercept + MTX Source: Klareskog L et al. Lancet 2004; 363: 675-81. Comparison of RA Studies % Achieving ACR Response MTX + TNF Antagonist in Established Disease 80 70 ACR20 Drug+MTX 60 ACR20 PBO+MTX 50 ACR50 Drug+MTX 40 30 ACR50 PBO+MTX 20 ACR70 Drug+MTX 10 ACR70 PBO+MTX 0 Inflix-10m g/kg/q8w Infliximab D2E7 40 q2w Adalimumab Etanercept 25 m g Etanercept 10 mg/kg/q4w 40 q2w 25 biw Lipsky Weinblatt Weinblatt NEJM 2000 A & R 2003 NEJM1999 biw Can We Predict Response to Abatacept, but not anti-TNF? Abatacept in TNF Inadequate Responders (ATTAIN) ACR20 ACR50 ACR70 60 % of patients 50.4 40 19.5 20.2 20 10.2 3.8 1.5 0 Placebo Genovese et al. N Engl J Med. 2005;353:1114. Abatacept 10mg/kg q 4w Can We Predict Risk of Serious Infection for RA Patients Treated with TNF Antagonists? Meta-analysis of Risk of Serious Infection from Adalimumab and Infliximab RCTs Bongartz, T. et al. JAMA 2006;295:2275-2285. Effect of HLA-DRB1 on ACR 50 Response Etanercept vs Methotrexate in the Early RA Etanercept Trial 80 70 60 50 40 2 copies SE 30 1 copy SE 20 0 copy SE 10 0 Methotrexate Etanercept 10 mg Etanercept 25 mg N=151 in the Etanercept 25 mg arm; OR (95% CI) for effect of SE = 4.3 (1.8 – 10.3) Criswell LA et al. Arthritis Rheum. 2004; 50(9): 2750-2756. HLA-DRB1 and Response to TNF Antagonists Summary of Published Literature Study Drug Outcome Criswell et al. (2004) Etanercept Positive association Kang et al (2005) Etanercept No association Marotte et al (2006) Infliximab No association Martinez et al (2004) Infliximab No association Miceli-Richard et al (2006) Adalimumab No association Effect of the TNF –308 G/A Polymorphism on Clinical Response to Infliximab (n=53) EULAR Good Response (Decrease of DAS-28 by 1.2) 100 80 60 80.5 71.7 40 41.7 20 0 Overall AA and A/G genotypes G/G genotype P=0.0086 Mugnier B. Arthritis Rheum. 2003; 48(7): 1849-1852 TNF Polymorphisms and Response to TNF Antagonists Summary of Published Literature Study Drug Outcome Mugnier et al. (2003) INF (N=59) Positive association Fonseca et al (2005) INF (N=22) Positive association Cuchacovich et al (2004) INF (N=22) No association Martinez et al (2004) INF (N=78) No association Padyukov et al (2003) ETA (N=123) Positive association* Criswell et al (2004) ETA (N=151) Positive association† * Combination of TNF and IL-10 SNPs. ** Extended Haplotype that included TNF/LTA and HLA-DRB1region Genetic Risk Factors are Stronger Predictors of Developing Infections (UTIs) than Clinical Risk Factors Etanercept vs Methotrexate in the Early RA Etanercept Trial Odds Ratio 95% CI Age >65 1.49 0.63 – 3.52 Elevated ESR 1.26 0.62 – 2.56 RF positive 1.32 0.43 – 4.06 Steroid use 1.15 0.58 – 2.27 ETA vs MTX 1.05 0.58 – 1.91 TNF –238 A 2.45 0.99 – 6.13 LTA +365 C 1.70 1.04 – 2.77 FcGR3a F 1.76 1.01 – 3.10 Hughes LB et al. Genes and Immunity 2004; 5: 641-647. Study Design and Analysis Issues in Pharmacogenetics SNP Selection for Pharmacogenetics • “Functional” SNP of candidate gene • High density genotyping of coding and non-coding SNPs of a specific candidate gene • SNP(s) of multiple genes in a metabolic pathway • Whole genome scan High Density Genotyping Project across the Whole Genome: Insights into the Correlation Structure of Alleles Insights from the HapMap Project and Related Studies Allele Frequencies of Drug Metabolizing Enzymes and Other Genes Vary across Different Population Groups CYP1A1 GSTM1 CYP2C19 DIA4 NAT2 CYP2D6 A. Bantu, Ethiopian, Afro-Caribbean B. Norwegians, Ashkenazi Jews, Armenians C. Chinese, New Guineans From Wilson, et al. Nature Genet 29:265-269, 2001 Relative power (%) Efficiency and power tag SNPs random SNPs ~300,000 tag SNPs needed to cover common variation in whole genome in CEU Average marker density (per kb) P.I.W. de Bakker et al. (2005) Nat Genet Advance Online Publication 23 Oct 2005 Can We “Tag” Candidate Inflammatory Gene SNPs Hypothesized to Modulate TNF Antagonist Response? Gene Common SNPs genotyped Related Haplotypes Haplotype Tag SNPs Haplotype Coverage by Tag SNPs TNF-α 9 4 4 87% IL-1β 20 4 3 100% IL-6 26 4 3 99% CTLA-4 11 12 5 100% 73 30 15 -- Future Directions Example of a diagnostic DNA microarray of polymorphisms of candidate genes to predict response and risk of toxicity (e.g. chemotherapeutic agent) Evans WE, Relling MV. Pharmacogenomics: Translating functional genomics into rational therapeutics. Science 286:487-491, 1999. Pharmacogenomic Studies May Be More Relevant to Predicting Response for Pleiotropic Drugs such as TNF Antagonists Roden, D. M. et. al. Ann Intern Med 2006;145:749-757 Gene Profiling in White Blood Cells Predicts Infliximab Responsiveness in Rheumatoid Arthritis • 33 RA patients treated with Infliximab • Responders vs nonresponders (decrease of DAS-28 1.2) • PBMC isolated from venous blood and total RNA extracted • mRNA collected at baseline hybridized to a microarray of 10,000 non-redundant cDNAs. • Real-time, quantitative reverse transcription PCR of selected mRNA also performed. • Statistical analysis included t- test and SAM (Significance Analysis of Microarrays) with a false discovery rate of <1%. Lequerre T et al. Arthritis Research and Therapy 2006: 8 R105 Gene Profiling in White Blood Cells Predicts Infliximab Responsiveness in Rheumatoid Arthritis Results • Overall 16/33 (48%) were EULAR responders. • The 33 patients randomly divided into: a) “Training” cohort (n=13) b) “Validation” cohort (n=20) • 41 mRNAs were differentially expressed in responders versus nonresponders s a function of the response to treatment • Differentially expressed genes were confirmed by qRT-PCR: – 20 transcript set – 8 transcript set Lequerre T et al. Arthritis Research and Therapy 2006: 8 R105 Gene Profiling in White Blood Cells Predicts Infliximab Responsiveness in Rheumatoid Arthritis 20 transcript set 8 transcript set Sensitivity 90 80 Specificity 70 100 PPV 75 100 NPV 88 83 The Future: Incorporate Biomarkers into Clinical Trials Evans and Johnson, Ann Rev Genom Hum Genet 29-39, 2001 Pharmacogenetics and “Personalized” Medicine Marsh, S. and McLeod HL. Hum. Mol. Genet. 2006 15:R89-93.