* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Metalloprotein wikipedia , lookup
Molecular ecology wikipedia , lookup
Peptide synthesis wikipedia , lookup
Non-coding DNA wikipedia , lookup
Proteolysis wikipedia , lookup
Multilocus sequence typing wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Biosynthesis wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Biochemistry wikipedia , lookup
Structural alignment wikipedia , lookup
Protein structure prediction wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
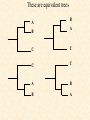
Progressive MSA • Do pair-wise alignment • Develop an evolutionary tree • Most closely related sequences are then aligned, then more distant are added. • Genetic distance - number of mismatched positions divided by the total number of matched positions (gaps not considered). Example • Domain: a segment of a protein that can fold to a 3D structure independent of other segments of the protein. • Card Domain • Caspase recruitment domains (CARDs) are modules of 90 - 100 amino acids involved in apoptosis signaling pathways. • http://www.mshri.on.ca/pawson/card.html These are equivalent trees A B B A C C C C A B B A QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Previous tree was Rooted These are Unrooted trees Gaps • Clustalw attempts to place gaps between conserved domains. • In known sequences, gaps are preferentially found between secondary structure elements (alpha helices, beta strands). Problem with Progressive Alignment: Errors made in early alignments are propagated throughout the MSA Profiles & Gaps • From an MSA, a conserved region identified and a scoring matrix (profile) constructed for that region. • Each position has a score associated with an amino acid substitution or gap. • Blocks- also extracted from MSA, but no gaps are permitted. • Block Server • Results – TLE short form – TLEl Long form Hidden Markov Models • Probabilistic model of a Multiple sequence alignment. • No indel penalties are needed • Experimentally derived information can be incorporated • Parameters are adjusted to represent observed variation. • Requires at least 20 sequences The bottom line of states are the main states (M) •These model the columns of the alignment The second row of diamond shaped states are called the insert states (I) •These are used to model the highly variable regions in the alignment. The top row or circles are delete states (D) •These are silent or null states because they do not match any residues, they simply allow the skipping over of main states. D1 D2 D3 D4 D5 D6 I4 I5 I6 I0 I1 I2 I3 B M1 M2 M3 M4 M5 M6 E The Evolution of a Sequence • Over long periods of time a sequence will acquire random mutations. – These mutations may result in a new amino acid at a given position, the deletion of an amino acid, or the introduction of a new one. – Over VERY long periods of time two sequences may diverge so much that their relationship can not see seen through the direct comparison of their sequences. Hidden Markov Models • Pair-wise methods rely on direct comparisons between two sequences. • In order to over come the differences in the sequences, a third sequence is introduced, which serves as an intermediate. • A high hit between the first and third sequences as well as a high hit between the second and third sequence, implies a relationship between the first and second sequences. Transitive relationship Introducing the HMM • The intermediate sequence is kind of like a missing link. • The intermediate sequence does not have to be a real sequence. • The intermediate sequence becomes the HMM. Introducing the HMM • The HMM is a mix of all the sequences that went into its making. • The score of a sequence against the HMM shows how well the HMM serves as an intermediate of the sequence. – How likely it is to be related to all the other sequences, which the HMM represents. Match State with no Indels MSGL MTNL B M1 M2 M3 M4 Arrow indicates transition probability. In this case 1 for each step E Match State with no Indels MSGL MTNL B M=1 S=0.5 T=0.5 M1 M2 M3 M4 E Also have probability of Residue at each positon Typically want to incorporate small probability for all other amino acids. MSGL MTNL B M=1 S=0.5 T=0.5 M1 M2 M3 M4 E Permit insertion states MS.GL MT.NL MSANI I0 I1 I2 I3 I4 B M1 M2 M3 M4 Transition probabilities may not be 1 E Permit insertion states MS..GL MT..NL MSA.NI MTARNL I0 I1 I2 I3 I4 B M1 M2 M3 M4 E MS..GL-MT..NLAG MSA.NIAG MTARNLAG DELETE PERMITS INCORPORATION OF LAST TWO SITES OF SEQ1 D1 D2 D3 D4 D5 D6 I4 I5 I6 I0 I1 I2 I3 B M1 M2 M3 M4 M5 M6 E The bottom line of states are the main states (M) •These model the columns of the alignment The second row of diamond shaped states are called the insert states (I) •These are used to model the highly variable regions in the alignment. The top row or circles are delete states (D) •These are silent or null states because they do not match any residues, they simply allow the skipping over of main states. D1 D2 D3 D4 D5 D6 I4 I5 I6 I0 I1 I2 I3 B M1 M2 M3 M4 M5 M6 E Dirichlet Mixtures • Additional information to expand potential amino acids in individual sites. • Observed frequency of amino acids seen in certain chemical environments – aromatic – acidic – basic – neutral – polar