* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Title goes here
Rheumatic fever wikipedia , lookup
Immune system wikipedia , lookup
Epoxygenase wikipedia , lookup
Adaptive immune system wikipedia , lookup
Gluten immunochemistry wikipedia , lookup
Polyclonal B cell response wikipedia , lookup
Antimicrobial peptides wikipedia , lookup
HLA A1-B8-DR3-DQ2 wikipedia , lookup
A30-Cw5-B18-DR3-DQ2 (HLA Haplotype) wikipedia , lookup
Molecular mimicry wikipedia , lookup
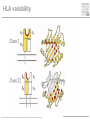
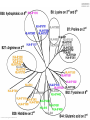
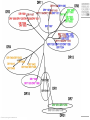
MHC Polymorphism MHC Class I pathway Figure by Eric A.J. Reits Expression of HLA is codominant Polymorphism and polygeny The MHC gene region http://www.ncbi.nlm.nih.gov/mhc/MHC.fcgi?cmd=init&user_id=0&probe_id=0&source_id=0&locus_id=0&locus_group=0&proto_id=0&banner=1&kit_id=0&graphview=0 Human Leukocyte antigen (HLA=MHC in humans) polymorphism - alleles http://www.anthonynolan.com/HIG/index.html HLA variability http://rheumb.bham.ac.uk/teaching/immunology/tutorials/mhc%20polymorphism.jpg MHC polymorphism •Selection pressure •Pathogens •Hosts •Cause of MHC polymorphism •Heterozygote advantage •Different MHC molecules bind different peptides •Heterozygous hosts have a broader immune response •Degree of MHC heterozygocity correlates with a delayed onset of progress to AIDS •Frequency-dependent selection by host-pathogen coevolution •Pathogens adapt to the most common MHC alleles •Rare alleles have a selective advantage Sets of MHC types •Each MHC molecule has a different specificity •If a vaccine needs to contain a unique peptide for each of these molecules it will need to comprise hundreds of peptides •Solution 1 •Select sets of a few HLA molecules that together have a broad distribution in the human population •Gulukota and DeLisi [1996] compiled lists with 3, 4, and 5 alleles which give the maximal coverage of different ethnic groups MHC Supertypes •Many of the different HLA molecules have similar specificities •HLA molecules with similar specificities can be grouped together •Methods to define supertypes •Structural similarities •Primary (sequence) •Tertiary (structure) •Shared peptide binding motifs •Identification of cross-reacting peptides •Ability to generate methods that can predict cross-binding peptides HLA polymorphism - supertypes •Each HLA molecule within a supertype essentially binds the same peptides •Nine major HLA class I supertypes have been defined •HLA-A1, A2, A3, A24,B7, B27, B44, B58, B62 Sette et al, Immunogenetics (1999) 50:201-212 HLA polymorphism - frequencies Supertypes Phenotype frequencies Caucasian Black Japanese Chinese Hispanic Average A2,A3, B7 83 % 86 % 88 % 88 % 86 % 86% +A1, A24, B44 100 % 98 % 100 % 100 % 99 % 99 % +B27, B58, B62 100 % 100 % 100 % 100 % 100 % 100 % Sette et al, Immunogenetics (1999) 50:201-212 HLA clustering method 1. Extract data from SYFPEITHI and MHCpep databases 2. Construct amino acid frequency vectors for each HLA molecule 3. Calculate distance between HLA molecules • The distance dij between two HLA molecules (i, j) is calculated as the sum over each position in the two motifs of one minus the normalized vector products of the amino acid’s frequency vectors (= cosine to the angle between the vectors) [Lyngsø et al., 1999]: 4. The distance matrices were used as input to the program neighbor from the PHYLIP package (http://evolution.genetics.washington.edu/phylip.html) O Lund et al., Immunogenetics. 2004 55:797-810 Logos of HLA-A alleles O Lund et al., Immunogenetics. 2004 55:797-810 Clustering of HLA alleles O Lund et al., Immunogenetics. 2004 55:797-810 Logos of HLA-B alleles O Lund et al., Immunogenetics. 2004 55:797-810 O Lund et al., Immunogenetics. 2004 55:797-810 Conclusions We suggest to – split some of the alleles in the A1 supertype into a new A26 supertype – split some of the alleles in the B27 supertype into a new B39 supertype. – the B8 alleles may define their own supertype – The specificities of the class II molecules can be clustered into nine classes, which only partly correspond to the serological classification O Lund et al., Immunogenetics. 2004 55:797-810 MHC class II pathway Figure by Eric A.J. Reits Virtual matrices HLA-DR molecules sharing the same pocket amino acid pattern, are assumed to have identical amino acid binding preferences. MHC Class II binding Virtual matrices – TEPITOPE: Hammer, J., Current Opinion in Immunology 7, 263-269, 1995, – PROPRED: Singh H, Raghava GP Bioinformatics 2001 Dec;17(12):1236-7 Web interface http://www.imtech.res.in/raghava/propred Prediction Results MHC class II Supertypes •5 alleles from the DQ locus (DQ1, DQ2, DQ3, DQ4, DQ5) cover 95% of most populations [Gulukota and DeLisi, 1996] •A number of HLA-DR types share overlapping peptide-binding repertoires [Southwood et al., 1998] Logos of HLA-DR alleles O Lund et al., Immunogenetics. 2004 55:797-810 O Lund et al., Immunogenetics. 2004 55:797-810