* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Chazin NMR Lecture - Center for Structural Biology
Franck–Condon principle wikipedia , lookup
Relativistic quantum mechanics wikipedia , lookup
Spin (physics) wikipedia , lookup
Scalar field theory wikipedia , lookup
Hydrogen atom wikipedia , lookup
Ferromagnetism wikipedia , lookup
Nuclear force wikipedia , lookup
Astronomical spectroscopy wikipedia , lookup
Rutherford backscattering spectrometry wikipedia , lookup
Molecular Hamiltonian wikipedia , lookup
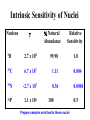
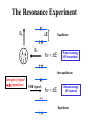
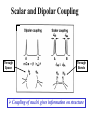
02/02/09 Biomolecular Nuclear Magnetic Resonance Spectroscopy BASIC CONCEPTS OF NMR • How does NMR work? • Resonance assignment • Structural parameters Reading: Chapter 22 in Protein and Peptide Drug Analysis “Solution Structure Determination of Proteins by NMR” Nuclear Spin • Nuclear spin angular momentum is a quantized property of the nucleus in each atom • The nuclear spin angular momentum of each atom is represented by a nuclear spin quantum number (I) • All nuclei with even mass numbers have I=0,1,2… • All nuclei with odd mass numbers have I=1/2,3/2... • NMR is possible with all nuclei except I=0 (e.g. 12C), but I=1/2 has simplest spin physics Biomolecular NMR primarily 1H, 13C, 15N (31P) Spin 1/2 Nuclei in a Magnetic Field Efficiency factornucleus Bo Energy DE = h g Bo Constants Strength of magnet Intrinsic Sensitivity of Nuclei Nucleus g % Natural Abundance Relative Sensitivity 1H 2.7 x 108 99.98 13C 6.7 x 107 1.11 0.004 15N -2.7 x 107 0.36 0.0004 31P 1.1 x 108 100 Prepare samples enriched in these nuclei 1.0 0.5 Variables Affecting Sensitivity Sensitivity (S) ~ D pop. (N vs N) S ~ DN = N = e-DE/kT N - DE is very small DN is small - DN ~ 1:105 (at room T) NMR has low sensitivity requires lots of sample! Efficiency factornucleus DE = h g Ho Constants Strength of magnet Increase sensitivity by increasing magnetic field strength or reducing electronic noise (cryo-probes) The Resonance Experiment DE Bo B1 hn = DE Equilibrium Pump in energy (RF transmitter) Non-equilibrium Strength of signal D (population) NMR signals hn = DE Release energy (RF receiver) Equilibrium NMR Terminology Chemical Shift & Linewidth The exact resonance frequency (chemical shift) is determined by the electronic environment of the nucleus Scalar and Dipolar Coupling Through Space Through Bonds Coupling of nuclei gives information on structure Resonance Assignment CH3-CH2-OH OH CH2 CH3 Which signal from which H atoms? Approach: use the scalar and dipolar couplings to match the set of signals with the molecular structure 2D NMR Spectra Facilitate Identification of Coupling F1 Coupled spins F2 Biomolecules Have Many Signals 1H NMR Spectrum of Ubiquitin ~75 residues, ~500 1H resonances Terminology: signals are overlapped Challenges For Using NMR to Study Biological Macromolecules • Hundreds-thousands of signals! • Must assign the specific signal for each atom • Thousands of couplings between nuclei- these also need to be assigned Critical Features of Protein NMR Spectra • Regions of the spectrum correspond to different parts of the amino acid • Tertiary structure leads to increased dispersion of resonances Regions of the 1H NMR Spectrum and Dispersion by the 3D Fold What would the unfolded protein look like? Critical Features of NMR Spectra of Biomolecules • Regions of the spectrum correspond to different parts of the amino acid • Tertiary structure leads to increased dispersion of resonances • Bio-macromolecules are polymers The nuclei are coupled to some (but not all!) other nuclei Spectra of Biomacromolecules: Overlapped Sub-Spectra *Groups of coupled nuclei* Each residue in the sequence gives rise to an independent NMR sub-spectrum, which makes the problems much simpler than if all spins were coupled Methods have been developed to extract each sub-spectrum from the whole Strategy to Assign Resonances in a Protein 1. Identify resonances for each residue (scalar) T G L S R G S 2. Put amino acids in order (dipolar) 1 2 3 4 5 6 7 R-G-S-T-L-G-S Same idea for any biopolymer (e.g. DNA, RNA) Even Sub-Spectra are Overlapped! 1H NMR Spectrum of Ubiquitin Resolve resonances by multi-dimensional experiments Use 2D NMR to Resolve Overlapping Signals 1D Sub-spectra overlapped 2D Crosspeaks resolved! Coupled spins Multi-Dimensional NMR Hb Ha HN If 2D cross peaks overlap go to 3D or 4D ….. Another Solution to the Overlap Challenge 1. Increase dimensionality of spectra to better resolve signals: 1D2D3D4D…. 2. Use hetero nuclei (13C,15N) to distinguish, coupling is efficient because only 1-bond t2 Hz HA t1 t3 Multi-Dimensional Heteronuclear NMR 15N-1H HSQC F1 Chemical Shift (15N) F2 Chemical Shift (1H) Advantages of Heteronuclear nD NMR Uses a second nucleus to resolve overlap of the first: chemical shift of each nucleus is sensitive to different factors More information to identify resonances Less sensitive to MW because this strategy uses large 1 and 2-bond scalar couplings