* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Nessun titolo diapositiva
Survey
Document related concepts
Unified neutral theory of biodiversity wikipedia , lookup
Occupancy–abundance relationship wikipedia , lookup
Habitat conservation wikipedia , lookup
Biodiversity action plan wikipedia , lookup
Latitudinal gradients in species diversity wikipedia , lookup
Introduced species wikipedia , lookup
Transcript
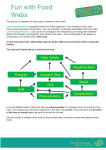
DYNAMICS OF COMPLEX SYSTEMS Self-similar phenomena and Networks Guido Caldarelli CNR-INFM Istituto dei Sistemi Complessi [email protected] 6/6 •STRUCTURE OF THE COURSE 1. SELF-SIMILARITY (ORIGIN AND NATURE OF POWER-LAWS) 2. GRAPH THEORY AND DATA 3. SOCIAL AND FINANCIAL NETWORKS 4. MODELS 5. INFORMATION TECHNOLOGY 6. BIOLOGY •STRUCTURE OF THE FIRST LECTURE 6.1) PROTEIN INTERACTION NETWORKS 6.2) FOOD WEBS 6.3) FOOD WEBS:Optimisation 6.4) PLANT TAXONOMIES •6.1 PROTEIN INTERACTION NETWORK Network of Interaction for the protein of Baker’s Yeast (Saccharomyces Cerevisiae) •6.1 PROTEIN INTERACTION NETWORK How do growth and preferential attachment apply to protein networks? • Growth: genes (that encode proteins) can be, sometimes, duplicated; mutations change some of the interactions with respect to the parent protein • Preferential attachment: the probability that a protein acquires a new connection is related to the probability that one of its neighbors is duplicated; proportional to its connectivity A. Vazquez et al., ComPlexUs 1, 38-44 (2003) •6.1 PROTEIN INTERACTION NETWORK The two hybrid method way of detecting protein interactions •6.1 PROTEIN INTERACTION NETWORK With the solvation free energies taken from an exponential probability distribution p(f) = e-f, we obtain P(k) ~ k-2 • The real network is random • The detection method sees only pairs with large enough binding constants • The binding constant is related to the solubilities of the two proteins • Solubilities are given according to some distribution •6.2 FOOD WEBS sequence of predation relations among different living species sharing the same physical space (Elton, 1927): Flow of matter and energy from prey to predator, in more and more complex forms; The species ultimately feed on the abiotic environment (light, water, chemicals); At each predation, almost 10% of the resources are transferred from prey to predator. •6.2 FOOD WEBS Set of interconnected food chains resulting in a much more complex topology: •6.2 FOOD WEBS: The topology Trophic Species: Set of species sharing the same set of preys and the same set of predators (food web aggregated food web). Trophic Level of a species: Minimum number of predations separating it from the environment. Basal Species: Species with no prey (B) Top Species: Species with no predators (T) Intermediate Species: Species with both prey and predators ( I ) Prey/Predator Ratio = BI IT •6.2 FOOD WEBS: The topology Pamlico Estuary (North Carolina): 14 species Aggregated Food Web of Little Rock Lake (Wisconsin)*: 182 species 93 trophic species How to characterize the topology of Food Webs? Graph Theory * See Neo Martinez Group at http://userwww.sfsu.edu/~webhead/lrl.html •6.2 FOOD WEBS: The degree irregular or scalefree? P(k) k- R.V. Solé, J.M. Montoya Proc. Royal Society Series B 268 2039 (2001) J.M. Montoya, R.V. Solé, Journal of Theor. Biology 214 405 (2002) •6.2 FOOD WEBS: The spanning tree A spanning tree of a connected directed graph is any of its connected directed subtrees with the same number of vertices. The same graph can have more spanning trees with different topologies. Since the peculiarity of the system (FOOD WEBS),some are more sensible than the others. •6.3 FOOD WEBS: The Optimisation 1 1 w XY 13 5 Out-component size: AX 1 1 AY 1 0,5 1 11 2 3 22 1 Sum of the sizes: CX 1 A Y Y X Allometric relations: Out-component size distribution P(A) : 0,6 1 5 8 Y nn X 1 10 33 CX CX A X 35 P(A) C C A C(A) 30 33 0,5 25 0,4 22 20 0,3 15 0,2 11 10 0,1 0,1 0,1 0,1 0,1 0,1 5 A 0 1 2 3 4 5 6 7 8 9 10 5 A 3 1 0 0 2 4 6 8 10 12 •6.3 FOOD WEBS: The Optimisation A0: metabolic rate B C0: blood volume ~ M Kleiber’s Law: B(M) M 3 / 4 C( A ) A General Case (tree-like transportation system embedded in a D-dimensional metric space): D1 the most efficient scaling is C( A ) A D West, G. B., Brown, J. H. & Enquist, B. J. Science 284, 1677-1679 (1999) Banavar, J. R., Maritan, A. & Rinaldo, A. Nature 399, 130-132 (1999). | 4 3 •6.3 FOOD WEBS: The Optimisation AX: drained area of point X Hack’s Law: C( A ) A L A0.6 3 2 •6.3 FOOD WEBS: The Optimisation •6.3 FOOD WEBS: The Optimisation (D.Garlaschelli, G. Caldarelli, L. Pietronero Nature 423 165 (2003)) •6.3 FOOD WEBS: The Optimisation Little Rock Webworld Little Rock Webworld S 182 182 S 93 93 L 2494 2338 L 1046 1037 B 0.346 0.30 B 0.13 0.15 I 0.648 0.68 I 0.86 0.84 T 0.005 0.02 T 0.01 0.01 Ratio 1.521 1.4 Ratio 1.14 1.16 lmax 3 3 lmax 3 3 C 0.38 0.40 C 0.54 0.54 D 2.15 2.00 D 1.89 1.89 1.11±0.03 1.12±0.01 1.15±0.02 1.13±0.01 2.05±0.08 2.00±0.01 1.68±0.12 1.80±0.01 Original Webs Aggregated Webs •6.3 FOOD WEBS: The Optimisation 1 0 1 0 C( A) A efficient C(A) A 1 2 P(A) A1 stable P(A) A 0 C( A) A 2 inefficient P(A) cost unstable •6.4 PLANT TAXONOMIES Lazio Utah Peruvian and Atacama Desert Amazonia Iran Argentina Ecosystem = Set of all living organisms and environmental properties of a restricted geographic area we focus our attention on plants in order to obtain a good universality of the results we have chosen a great variety of climatic environments •6.4 PLANT TAXONOMIES Linnean Tree = hierarchical structure organized on different levels, called taxonomic levels, representing: • • classification and identification of different plants history of the evolution of different species phylum subphylum class subclass order A Linnean tree already has the topological structure of a tree graph family genus species • each node in the graph represents a different taxa (specie, genus, family, and so on). All nodes are organized on levels representing the taxonomic one • all link are up-down directed and each one represents the belonging of a taxon to the relative upper level taxon Connected graph without loops or double-linked nodes •6.4 PLANT TAXONOMIES P(k) Degree distribution: k P ( k ) k ~ 2.5 0.2 The best results for the exponent value are given by ecosystems with greater number of species. For smaller networks its value can increase reaching = 2.8 - 2.9. •6.4 PLANT TAXONOMIES Colosseo (Terza Università, Rome) 6 historical periods (1643 - 2001) historical events climatic changes Valmalenco (Bernina) (University of Pavia) (G. Rossi, M. Gandini) 3 historical periods (1949 - 2003) climatic changes (Global Warming) •6.4 TAXONOMY: Real Subsets Tiber Mte Testaccio Aniene Lazio k =2.52 0.08 City of Rome Colli Prenestini k =2.58 0.08 k 2.6 ≤ ≤ 2.8 •6.4 TAXONOMY: Random Subsets P(k) P(k) P(k) random extraction of 100, 200 and 400 species between those belonging to the big ecosystems and reconstruction of the phylogenetic tree ROME LAZIO k P(k) P(k) • Simulation: k k P(k)=k -2.6 k k •6.4 TAXONOMY: Random Subsets P(kf, kg) that a genus with degree kg belongs to a family with degree kf kf=1 kf=2 kf=3 kf=4 kg = ∑g kg P(kf,kg) fixed P(kf,kg) kg - fixed ~ 2.2 0.2 kf kg P(ko,kf) that a family with degree kf belongs to an order ko=1 ko=2 ko=3 ko=4 P(ko,kf) kf kf = ∑f kf P(ko,kf) fixed - fixed ~ 1.8 0.2 kf with degree ko ko •6.4 TAXONOMY: Random Subsets 1) create N species to build up an ecosystem 2) Group the different species in genus, the genus in families, then families in orders and so on realizing a Linnean tree - Each species is represented by a string with 40 characters representing 40 properties which identify the single species (genes); - Each character is chosen between 94 possibilities: all the characters and symbols that in the ASCII code are associated to numbers from 33 to 126: P g H C ) %o r ? L 8 e s / C c W & I y 4 ! t G j z AB 4 2£ ) k , ! d q 2= m: f V Two species are grouped in the same genus according to the extended Hamming distance dWH: c1i = character of species 1 c2i = character of species 2 ba Z with i=1,……….,40 with i=1,……….,40 dEH = ( ∑i=1,40 |c1i - c2i| )/40 •6.4 TAXONOMY: Random Subsets P g H C ) %o r ? L G j 4 2£ ) k , ! d dEH ≤ C |c1i - c2i| = 17 species 1 same genus Fixed threshold species 2 genus = average of all species belonging to it c14 c(g)4 P g H C ) %o r ? L ( c1i + c2i )/2 G j : 4 2£ ) k , ! d c24 Same proceedings at all levels with a fixed threshold for each one At the last level (8) same phylum for all species (source node) •6.4 TAXONOMY: Random Subsets No correlation: species randomly created with no relationship between them Genetic correlation: species are no more independent but descend from the same ancestor • No correlation: ecosystems of 3000 species each character of each string is chosen at random quite big distance between two different species: P(k) dEH ~ 20 (S . ` U d ~j <@a ~N f K Mg X w´ * : * 4 " j ° z G 9 / F y 2 J ´ R _ x 5 K L ` < G ´ D Q b mV U W ; d L U x o g Z k * 8 y u N v D K Z + { C x 6 I 6 d z k (top ~ 1.7 0.2 bottom ~ 3.0 0.2 ) •6.4 TAXONOMY: Random Subsets single species ancestor of all species in the ecosystem at each time step t a new species appear: - chose (randomly) one species already present in the ecosystem - change one of its character 3000 time steps Environment = average of all species present in the natural selection the ecosystem at each time step t. At each time step t we calculate the distance between the environment and each species: dEH < Csel survival dEH > Csel extinction small distance between different species: dEH ~ 0.5 g 5 0 _ " & y = E o [ l R C ( x z G ? g = X %W @ @ / X r ] T K g ? 6 Y G ^ Q z g 5 0 _ " & y = E o [ : R C ( x z G ? 0 = / %W ´ S / X r ] T K g ? 6 K ^ ^ Q z P(k) ~ k - k ~ 2.8 0.2 •6.4 TAXONOMY: Random Subsets Correlated: k k P(k) Not Correlated: •CONCLUSIONS Results: networks (SCALE-FREE OR NOT) allow to detect universality (same statistical properties) for FOOD WEBS and TAXONOMY. Regardless the different number of species and environment STATIC AND DYNAMICAL NETWORK PROPERTIES other than the degree distribution allow to validate models. NEITHER RANDOM GRAPH NOR BARABASI-ALBERT WORK Future: models can be improved with particular attention to environment and natural selection FOR FOOD WEBS AND TAXONOMY new data COSIN COevolution and Self-organisation In dynamical Networks RTD Shared Cost Contract IST-2001-33555 http://www.cosin.org • • • • • Nodes Period of Activity: Budget: Persons financed: Human resources: EU countries Non EU countries EU COSIN participant Non EU COSIN participant 6 in 5 countries April 2002-April 2005 1.256 M€ 8-10 researchers 371.5 Persons/months