* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Root proteases: reinforced links between nitrogen uptake and
Survey
Document related concepts
Cultivated plant taxonomy wikipedia , lookup
Arabidopsis thaliana wikipedia , lookup
Historia Plantarum (Theophrastus) wikipedia , lookup
Indigenous horticulture wikipedia , lookup
History of botany wikipedia , lookup
Ornamental bulbous plant wikipedia , lookup
Plant use of endophytic fungi in defense wikipedia , lookup
Venus flytrap wikipedia , lookup
Plant defense against herbivory wikipedia , lookup
Plant secondary metabolism wikipedia , lookup
Plant physiology wikipedia , lookup
Plant morphology wikipedia , lookup
Plant stress measurement wikipedia , lookup
Hydroponics wikipedia , lookup
Transcript
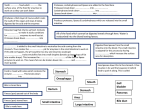
Physiologia Plantarum 2012 Copyright © Physiologia Plantarum 2012, ISSN 0031-9317 Root proteases: reinforced links between nitrogen uptake and mobilization and drought tolerance Ajay Kohlia,∗ , Joan Onate Narcisoa , Berta Mirob and Manish Raoranea a b Plant Breeding, Genetics, and Biotechnology Division, International Rice Research Institute, DAPO, Metro Manila, Philippines Crop and Environmental Sciences Division, International Rice Research Institute, DAPO Box 7777, Metro Manila, Philippines Correspondence *Corresponding author, e-mail: [email protected] Received 22 June 2011; revised 15 November 2011 doi:10.1111/j.1399-3054.2012.01573.x Integral subcellular and cellular functions ranging from gene expression, protein targeting and nutrient supply to cell differentiation and cell death require proteases. Plants have unique organelles such as chloroplasts composed of unique proteins that carry out the unique process of photosynthesis. Hence, along with proteases common across kingdoms, plants contain unique proteases. Improved knowledge on proteases can lead to a better understanding of plant development, differentiation and death. Because of their importance in multiple processes, plant proteases are actively studied. However, root proteases specifically are not as well studied. The associated rhizosphere, organic matter and/or inorganic matter make roots a difficult system. Yet recent research conclusively demonstrated the occurrence of endocytosis of proteins, peptides and even microbes by root cells, which, hitherto known for specialized pathogenesis or symbiosis, was unsuspected for nutrient uptake. These results reinforced the importance of root proteases in endocytosis or root exudate-mediated nutrient uptake. Rhizoplane, rhizosphere or in planta protease action on proteins, peptides and microbes generates sources of nitrogen, especially during abiotic stresses such as drought. This article highlights the recent research on root proteases for nitrogen uptake and the connection of the two to drought-tolerance mechanisms. Drought-induced proteases in rice roots, as known from rice expression databases, are discussed for future research on certain M50, Deg, FtsH, AMSH and deubiquitination proteases. The recent emphasis on linking drought and plant hydraulics to nutrient metabolism is illustrated and connected to the value of a systematic study of root proteases in crop improvement. Abbreviations – AA, amino acid; ABA, abscissic acid; AMSH, associated molecule with the SH3 domain of STAM; ATP, adenosine triphosphate; BNF, biological nitrogen fixation; C, carbon; ClpP1, caseinolytic protease P1; CP, cysteine protease; Deg, degradation of periplasmic protein; ER, endoplasmic reticulum; FC, field capacity; FtsH, filamentation temperature-sensitive H; GFP, green fluorescent protein; IPT, isopentenyltransferase; LHT, lysine histidine transporter; MT, membrane-tethered; N, nitrogen; OAA, oxaloacetate; OTU, ovarian tumor; PAGE, polyacrylamide gel electrophoresis; PDZ, post synaptic density protein (PSD95), drosophila disc large tumor suppressor (DlgA) and zonula occludens-1 protein (zo-1); PSII, photosystem II; PTR, peptide transporter; RIP, regulated intramembrane proteolysis; SREBP, sterol-regulated element binding protein; SUMO, small ubiquitin-related modifier; TF, transcription factor; Ulp, ubiquitin-like protease; UPR, unfolded protein response. Physiol. Plant. 2012 Introduction Proteases cleave peptide bonds, leading to protein breakdown. Thus, the general perception on biological functions of proteases was largely restricted to degradation of toxic, undesirable and damaged proteins, releasing amino acids (AAs) that could be reused in protein synthesis. However, proteases are now known to regulate various biological processes spanning the life cycle of plants, starting with cell division to specialized tissue, organ or organelle formation and development such as gametes, embryo, seed coat, stomata and chloroplast (Van der Hoorn 2008). Their activity in response to external stimuli such as pest and pathogen attack (Xia 2004, Baek and Choi 2008) or in response to internal developmental stimuli such as senescence has also been characterized (Woltering 2004). Sequenced genomes of Arabidopsis and rice reveal close to 700 and 800 proteases, respectively, which are classified into clans and families, while members within the families also exhibit functional variation (Van der Hoorn 2008). Plants with larger genomes are likely to have a higher number of proteases. However, the biological function of only around 40 proteases is known. Considering the involvement of proteases in various aspects of plant growth, development, survival and senescence, a better understanding of their phenotypic effects and molecular mechanisms is imperative. This is especially true for plant-specific members of some families of proteases such as the plastidial proteases. For example, chloroplast biogenesis and function involve the chloroplast genome-encoded caseinolytic protease P1 (ClpP1) whose mutation arrests etiolated shoot development in tobacco plants (Kuroda and Maliga 2003). Nuclear-encoded Clp proteases could not complement ClpP1 mutation, thus suggesting that ClpP1 performs more than the housekeeping protein breakdown function. Kuroda and Maliga (2003) speculated that ClpP1 may act on a specific regulatory protein. Similarly, the plastidial filamentation temperature-sensitive H (FtsH) protease complex is involved in the critical function of turnover of the photosystem II (PSII) reaction center D1 protein. It is also involved in other processes required for the development and maintenance of the photosynthetic apparatus (Liu et al. 2010). Studies on plant proteases have advanced substantially over the past few years and an ever-increasing understanding of the role of proteases in the life cycle of plants is emerging. The regulatory role of plant proteases in post-translational modification of proteins by limited proteolysis at specific sites is being recognized as a key route to involvement in different functions. Limited and site-specific proteolysis gives rise to functional enzymes and regulatory proteins and peptides, facilitates subcellular targeting, and is required for protein assembly (Schaller 2004). In a recent review, Van der Hoorn (2008) collated information on 40 plant proteases with known phenotypes and described some of the well-characterized proteases representing different functional aspects in different cellular processes. Only two of the plant proteases described were concerned with roots. One affected root meristem maintenance in Arabidopsis (Casamitjana-Martinez et al. 2003) and the other had a role in Medicago truncatula root infection by Sinorhizobium meliloti (Combier et al. 2007). In the last 3 years, only one more protease in roots has since been assigned a phenotype, a cysteine protease (CP) that is a negative regulator of nodules and bacteroid senescence in Astragalus sinicus (Li et al. 2008). A relative lack of detailed characterization of root proteases is perhaps indicative of the difficulties associated with working with roots under natural environments. Molecular and genetic studies on roots have largely concentrated on root development, morphology and architecture (reviewed by Rost 2011). The other popularly explored aspect of roots is the root–microbe symbiosis for nitrogen fixation. Murray (2011) and Yokota and Hayashi (2011) reviewed the genes involved in root–microbe symbiosis in legumes. Traditionally, roots have been considered little more than a conduit for water and dissolved minerals. Genetic controls and molecular biology of mineral acquisition by roots, including both macronutrients such as nitrogen, phosphorus, potassium, sulfur and calcium and micronutrients such as iron, molybdenum, boron, copper, manganese, zinc, chloride, silicon, sodium and nickel, etc., are now being elucidated. Kraiser et al. (2011) presented an integrated view on different mechanisms that help plants to acquire and maintain an adequate nitrogen supply. These included root-specific gene expression and function modulation, root growth and development modulation and root–microbe association modulation. Similarly, the state of knowledge on root uptake and use of phosphorus (Panigrahy et al. 2009), potassium (Szczerba et al. 2009) and sulfate (Rouached et al. 2009) has also been reviewed, while Chen et al. (2008) reviewed membrane transporters involved in the uptake of the three macronutrients, i.e. nitrogen, phosphate and potassium. The three main aspects of root research – root development and symbiotic and non-symbiotic nutrient acquisition – were reviewed for an integrated understanding of root adaptations to biotic and abiotic constraints on root function and plant health (Hodge et al. 2009). Such integrative studies on nutrient acquisition are increasing due to an imperative need to optimize nutrient application, uptake and use as an important aspect of precision Physiol. Plant. 2012 farming in the future so as to increase land and plant yield potential under normal and/or stress conditions. Improving the yields of major cereal crops is a key to future food security under population and climatic pressure. As cereals do not depend on nutrient availability through symbiosis, an improved understanding of nonsymbiotic nutrient uptake and mobilization is desirable. Interestingly, recent reports on two modes of nutrient uptake have added to our knowledge on the mechanisms that roots employ for the purpose. One is root exudate-mediated nutrient uptake, which was known for some time, but more details of exudate components and their roles are emerging now. The second is endocytosis of proteins and intact bacteria by root cells. Endocytosis by roots was known in connection with symbiotic or pathogenic relationships, but clear demonstrations of the process being used for nutrient acquisition have been provided recently. Both mechanisms invoke a crucial role for root proteases, exuded or internal, in order to break down, use and remobilize the scavenged AAs, peptides and proteins and thus maintain the nitrogen status of plants. The nitrogen status of plants under stress is a critical factor in stress tolerance. Under water-deficit conditions, for example, nitrogen content can decline drastically (Gonzalez-Dugo et al. 2010). A corollary to this is that increased root proteases would have a critical role in stress tolerance because they can mobilize nitrogen from external or internal sources. This is somewhat a different view on the role of plant proteases under stress, specifically wheat CPs, whereby an increase in their stress-associated expression in leaves was proposed as a marker of susceptibility to water-deficit (Simova-Stoilova et al. 2010). A similar study conducted on the roots of red clover did not show an increase in CPs till after 21 days of stress, after which the roots were terminally consigned to senescence (Webb et al. 2010). Nevertheless, preliminary results from our laboratory combined with extensive information gleaned from rice expression databases clearly implicate a role for rice root proteases in compensating for the nitrogen stress concomitant to drought. It is argued that proteases that are seen to be specifically upregulated in roots under drought belong to two categories, cell death and nutrient mobilization/regulation. It is further suggested that during drought the balance may shift more toward the latter category in tolerant accessions of rice, conferring a better ability to uptake and remobilize nitrogen under stress. Such a shift from vacuolar degradative protease activity to nonvacuolar energy-dependent regulatory protease activity was shown in wheat leaves in relation to developmental stage and water saturation deficit (Wisniewski and Zagdanska 2001), although total protease activity was not Physiol. Plant. 2012 substantially different. The importance of this shift under stress was not established and such observations have not been made in roots yet. We suggest that further research on stress-responsive proteases in roots and their specific roles would add valuable information on integrating the root–leaf and/or root–tiller response in terms of hydraulics and nutrient re-mobilization. This would be particularly useful for rice, whose roots seem to survive long after the aerial parts have wilted. The return of favorable conditions supports a fast-tracked growth of new tillers that could bear seeds. This is reminiscent of red clover roots surviving for a long time after defoliation or grazing (Bingham and Rees 2008, Webb et al. 2010). Root proteases and nitrogen uptake The root–soil interface is an active site for the exchange of organic and inorganic C and N, the two major elements of plant nutrition, whereas other macroand micronutrients are also sourced from the rhizosphere–soil continuum. The C and N exchange at the roots driven by variables of soil, microbial biomass and plants is highly complex and is spatiotemporally regulated. Jones et al. (2009) presented four possible utilities for such exchanges, two of which centered on root–root and root–microbe interactions and the other two on nutrient uptake and distribution by plants. For root–microbe interaction between Medicago and Sinorhizobiummeliloti, peptidases were found to be an important component of the root exudates (De-la-Pena and Vivanco 2010). Interestingly, however, Godlewski and Adamczyk (2007) found that proteases were secreted by the roots of 15 different agricultural and wild plant species without any microbial or other artificial means of inducing protease secretion. Their studies with wheat (Adamczyk et al. 2008) and Allium (Adamczyk et al. 2009) suggested that such protease secretion was part of the plants’ strategy to increase free AAs as a source of N. Thus, AAs in soil could result from the activity of such exuded root proteases on proteins and peptides outside of the roots. Alternately, increased root protease activity within the roots under stress could be the source of AAs in the soil, again as part of the root exudate. Indeed, it was established that increased root exudates of Festuca rubra under stress were richer in AAs (Paterson et al. 2003), while Lesuffleur et al. (2007) reported that root exudates of different plants under stress were rich in different AAs. These and other studies (Schobert and Komor 1987, Persson and Nasholm 2002) lend credibility to the proposition by Jones et al. (2009) that one possible utility of exudates is re-absorption of some of their components by roots to maintain C and N balance. That roots do uptake AAs from soil and use them in vivo was demonstrated through a combined proteomic and isotope tracer approach, whereby labeled glycine supplied as an N source was traced in plant metabolites in Lolium perenne (Thornton et al. 2007). In this study, root proteome analysis with and without glycine exhibited upregulation of a glycine metabolism enzyme, among others, in the former case. The proteases in the rhizosphere may equally come from microbes. Vágnerová and Macura (1974) demonstrated that protease activity because of microbes was higher in the wheat rhizoplane than in the rhizosphere and surrounding soil. However, plant root exudates exercise a selective influence so that only a small and specific proportion of microbial diversity can inhabit the rhizosphere and/or the rhizoplane, and the rhizosphere inhabitants may vary depending also on environmental and plant growth variables (Hartmann et al. 2009). Thus, proteases in root exudates or within the roots or those from rhizoplane non-symbiotic bacteria may be responsible for protein breakdown, facilitating N uptake in the form of AAs. Indeed, a root–microbe symbiotic association is another mechanism of organic N availability to plants. Although there was evidence that plants could use AA from soil or culture media, these were artificially supplied. Paungfoo-Lonhienne et al. (2008) demonstrated that axenically cultivated non-symbiotic Hakeaactites (a woody plant) as well as the herbaceous Arabidopsis could use protein as a source of nitrogen in the absence of any other symbiotic or non-symbiotic organism. Their results validated a role for root proteases in the absence of any other microbe, when the root surface fluoresced from proteolysis of an externally supplied protein–chromophore complex capable of fluorescence only upon proteolysis. More importantly, it was also shown with the use of green fluorescent protein (GFP) and through the lack of protein degradation products in the incubation media that intact protein was taken up by the roots as long as root hair were present. The same group further elegantly demonstrated that roots of tomato and Arabidopsis were capable of taking up intact Escherichia coli and yeast cells, which were then degraded within the root cells and were at least a source of nitrogen, as detected by an increase in 15 N concentrations in the leaves after incubation of tomato roots with 15 N-labeled E. coli (Paungfoo-Lonhienne et al. 2010). Such consumption of E. coli N clearly implicated root proteases. There was evidence through induction of specific genes and structures that the process of taking up intact microbes by roots involves root outgrowths that surround E. coli cells stuck to the root surface through mucilage secreted either from the roots or from the bacteria. Endocytosis was also implicated as a possible process that could facilitate microbe uptake because induction of genes for cytoskeleton structure and re-organization was noticed. The importance of endocytosis in different aspects of plant–microbe interactions, microbe recognition and colonization of plant cells by endosymbionts has been recently reviewed with a special emphasis on clathrin-mediated endocytosis (Leborgne-Castel et al. 2010). These results clearly linking N uptake and use to root proteases, apart from the uptake of inorganic N or symbiotic N in plants, are from studies over the past 4 years only on a limited number of species. Although not much information is available on any such parallels in crop plants in field conditions, the stage is set for such a search. Drought and nitrogen uptake The importance of the relationship between drought and N uptake by plants is becoming steadily clear. Drought invariably reduces biological nitrogen fixation (BNF) and consequently the N content, biomass and yield of plants. In peanut, BNF and biomass increasingly declined under 2/3 of field capacity (FC) water and 1/3 of FC for 12 different lines (Pimratch et al. 2008). However, there was a positive correlation between BNF and biomass under both water-deficit conditions and the relationship was stronger under severe water-deficit. The results suggested that maintenance of high BNF under drought would be useful in maintaining high yield under waterdeficit conditions. Indeed, this approach was earlier used to select Glycine max genotypes with high ability to fix N under drought, which led to the release of drought-tolerant varieties (Chen et al. 2007). However, it is not only the symbiosis-dependent plants that cope better with drought under improved N content, but also the symbiosis-independent plants. For example, in rice, various physiological parameters such as photosynthesis, relative chlorophyll content, water use efficiency, transpiration and overall growth were significantly higher with the application of N during drought than under drought with no N application (Suralta 2010). Similarly, under drought stress applied to barley at three growth stages, tillering, shooting and earing, the negative effect on grain yield could be relatively relieved with N fertilizer application (Krcek et al. 2008). Reviewing water-deficit and nitrogen nutrition of crops, Gonzalez-Dugo et al (2010) elaborated that mineral N from the rhizosphere is largely made available through the transpiration stream and that under waterdeficit mineral N fluxes decrease in the xylem. The concomitant N assimilation is also more sensitive to water scarcity. Why does increased N, organic or Physiol. Plant. 2012 inorganic, through transpiration pull or through exudates and/or endocytosis, help to relieve the effects of drought? Plants under stress tend to maintain the internal nutrient status of the C–N ratio either by modulating the partitioning of nutrients or by nutrient re-mobilization. Vegetative growth may be controlled to limit the tissues that require balanced nutrient status. For example, Norway spruce under drought but with an N source available exhibited less new shoot development than this species under drought, but with no N source available (Rosengren-Brinck and Nihlgard 1995). This suggested preferential allocation of available N to maintain the nutrient status of a limited body of the plant. In rice, reproductive tiller formation may be favored over new leaf development under drought and with N availability. Under progressive drought, an imbalance in the internal nutrient status of aerial parts is more likely to occur before terminal water-deficit per se. This is because the perception of water-deficit initiates hydraulic and hormonal changes that restrict stomatal opening, in turn restricting transpiration pull and carbon dioxide entry, both of which initiate changes in C and N metabolism and assimilation. In tomato, root restriction stress, which mimics water stress, affected xylem sap ABA concentration, which in turn directly affected aerial characteristics such as stomatal conductance, leaf water potential, intercellular CO2 concentration, accumulation of carbohydrates, maximum carboxylation rate of Rubisco, ribulose-1,5-bisphosphate regeneration and quantum yield of PSII electron transport (Shi et al. 2008). Hence, under drought, along with setting in motion the physiological and molecular machinery for efficient water uptake and use, the plant’s effort is also toward activating mechanisms that ensure efficient nutrient uptake, re-mobilization and use. For nutrient uptake and mobilization by roots the activity of exuded or internal root may be critical. Proteases in the aerial parts may also be activated for the purpose of leaf N re-mobilization. However, root proteases are likely to play a more important role as being at the site of N uptake. Our preliminary results discussed below demonstrate much higher protease activity in the roots of different rice genotypes than in the shoots, both with and without drought. Relation between root proteases, nutrient status and drought Between ammonium and nitrate as an N source under drought, rice showed a preference for the former. A direct link with increased photosynthesis in the presence of ammonium nutrition was noticed (Guo et al. 2007). Rice is special in that flooded field and drought situation Physiol. Plant. 2012 change the main source of N from ammonium to nitrate, as also the degree of non-symbiotic BNF, in turn influencing the relative importance of ammonium and nitrate (Buresh et al. 2008). However, uptake of AAs by plant roots as additional or alternate source of N through AA transporters has been known (Nasholm et al. 2009). Interestingly, it was shown by carbon and nitrogen isotope labelling that wheat roots took up AAs in the form of tripeptides in preference to nitrates and single AAs. Uptake of nitrates was inferior to that of ammonium, L-trialanine and even the biologically unutilized D-trialanine (Hill et al. 2011). Also, as illustrated above, peptides, proteins and/or microbes of the rhizosphere/rhizoplane could be used as a source of N under drought and root proteases may have an important role in hydrolyzing soil proteins or in digesting those taken up by roots. Drought-induced proteases are known from the aerial parts of plants such as Arabidopsis (Koizumi et al. 1993), pea (Jones and Mullet 1995), Phaseolus (Hieng et al. 2004) and wheat (Zagdanska and Wisniewski 1996, 1998, Demirevska et al. 2008, Zang et al. 2010). Such induction correlates positively with drought-sensitive or drought-tolerant cultivars. This confusion exists if detailed characterization of the proteases involved is not available because senescence-related proteases as well as nutrient re-mobilization and regulatory proteases both can be induced under drought. It is possible that the former class of proteases is induced more in droughtsensitive genotypes while the latter class is induced more in tolerant genotypes. For example, increased protease induction in drought-sensitive genotypes was noticed in Phaseolus (Roy-Macauley et al. 1992, Hieng et al. 2004), while in wheat the drought-tolerant varieties exhibited increased induction (Zagdanska and Wisnievski 1998, Demirevska et al. 2008). Importantly, the proteases studied in wheat were ATP-dependent regulatory proteases while in Phaseolus they were vacuolar serine and/or CPs. For wheat under water-deficit, total protease activity increased and CPs largely contributed to the increase (Zagdanska and Wisnievski 1996). While Simova-Stoilova et al. (2006) associated increased CP activity with sensitive wheat genotypes, Zang et al. (2010) recently associated another wheat CP (TaCP) with drought tolerance through transgenic validation in Arabidopsis, whereby constitutive expression of TaCP enhanced the drought tolerance of the transgenic Arabidopsis. Indeed, different CPs are associated with senescence and drought (Khanna-Chopra et al. 1999) and both categories, degradative and processive CPs, can be upregulated. Depending on which CPs were characterized, the association with tolerant or susceptible genotypes could vary. Despite evidence for the importance of intracellular proteolysis during drought, most likely in re-organizing cellular metabolism, the role of specific proteases under stress remains uncertain. This is even more the case for root proteases. The limited information available on root proteases is mostly from work on isolated root tips and largely in relation to C starvation. For example, a root protease upregulated in maize root tips under glucose starvation was described (Chevalier et al. 1995) and it was shown that protein catabolism in maize root tips followed fatty acid catabolism under C starvation (Dieuaide-Noubhani et al. 1997). In general, endopeptidase and protease expression increased in excised maize root tips under C-starvation conditions (Brouquisse et al. 2001, 2007). Sugar starvation in tomato roots led to a reduction in root carbohydrate as well as in root protein content (Devaux et al. 2003) implicating increased protease activity in the latter case. In the case of intact roots of Phaseolus vulgaris under stress, protease activity did not increase in the first 3 days of stress and AA levels decreased, including that of asparagine (Bathellier et al. 2009). However, intact roots within 3 days of stress may have still not reached a point of critical stress more easily achieved in excised roots as exemplified by the increase in AAs after excision of red clover roots from shoots (Bingham and Rees 2008). Interestingly, asparagine synthetase – the enzyme for synthesis of asparagine, the major N transport molecule in plants during stress – was highly upregulated in Arabidopsis under sugar stress imposed by darkness (Gaufichon et al. 2010). This suggests that during C starvation the plant prepares for N re-mobilization. What is the relationship between drought, C starvation and protease upregulation? The water deficit-mediated hydraulic signal from roots to shoots results in changes in leaf cell turgor leading to hormonal signaling-mediated stomatal closure (Christmann et al. 2007). Stomatal closure is useful in limiting transpiration under drought, but it also limits carbon dioxide intake, which in turn affects net photosynthesis (McDowell et al. 2008, Pinheiro and Chaves 2011). A change in photosynthesis leads to the initiation of drought-induced C starvation, which results in a sugar-mediated signaling cascade for the re-organization of cellular metabolism. Overall, droughtinduced C starvation is seen to be the major threat to plant survival concomitant to changes in C–N ratio and in plant hydraulics (McDowell, 2011). Recent findings suggest that a number of late molecular responses are more to changes in carbohydrate availability than to environmental cues (Smith and Stitt 2007) and that plants may survive some period of complete loss of conductivity (McDowell 2011). Additionally, during drought, nutrient and mineral uptake decline (Sardans and Penuelas 2005), transport to the aerial parts decreases (Hu and Schmidhalter 2005) and energy costs of converting nitrogen into usable forms increase (Farooq et al. 2009, Pinheiro and Chaves 2011). Under such nitrogendeficient conditions, the action of proteases may supply the AAs, such as asparagine and glutamine for N re-mobilization. Additionally, root exudate-mediated and/or root endocytosis-mediated uptake of water, nutrients, organic molecules and microbes can provide a lifeline to the plant, depending on the timing and severity of drought. It is to be noted that the organic molecules sourced through the activity of proteases are not just N sources, but also C sources as observed with asparagine breakdown into oxaloacetate (OAA) through the activity of the amidohydrolase asparaginase. The OAA can be used in the Krebs cycle, which produces the reducing power as hydrogen atoms for the electron transport chain and intermediates that can be further anabolized (Simpson 2011). Additional root growth observed under nutrient deficiency was proposed to be important in sourcing additional soil area for nutrient uptake (LopezBucio et al. 2003). Hence, drought-induced root growth is opposite to shoot growth retardation under drought. Nutrients for root growth are sourced as organic matter from the soil, thus saving energy in converting inorganic N into an AA (Nasholm et al. 2009). Another source of water and nutrients is the process of autophagy. Cells respond to stress with a burst of autophagic activity through co-ordinated upregulation of autophagy genes at the onset of starvation in Arabidopsis and proteases are part of the autophagic gene complement (Rose et al. 2006). A lack of these proteases makes Arabidopsis mutants hypersensitive to C and N starvation (Thompson et al. 2005). Indeed, root tips of Arabidopsis undergoing water stress exhibited autophagy (Duan et al. 2010). During drought, nutrients and even water sourced through different mechanisms by roots, either from outside or within, need to be transported to the aerial parts. The uptake of AAs and their transfer into the root xylem have been described in Ricinus communis (Schobert and Komor 1990), and the corresponding AA transporters in this plant have been characterized (Bick et al. 1998). Overall importance of organic N (AAs, peptides and proteins) uptake by roots and associated transporters were reviewed by Nasholm et al. (2009) and Tegeder and Rentsch (2010). Nitrogenous nutrients in xylem can flow to phloem and to the shoots (Atkins 2000, Zhang et al. 2010) through specific transporters, including peptide transporters (PTRs) (Lubkowitz 2011). Proton-coupled, independent or pH-gradientdriven AA and sugar influx, efflux and bidirectional transporters at the plasma membrane and tonoplast have been described (Yang et al. 2010, Okumoto and Physiol. Plant. 2012 Pilot 2011). The lysine histidine transporter (LHT1) is a high affinity broad-spectrum AA transporter expressed in the leaf and root epidermis and it directly affects AA uptake and distribution in plants (Hirner et al. 2006). Upregulation of LHT1 was recently shown in drought-tolerant rice plants generated by overexpression of the cytokinin synthesis gene, isopentenyltransferase (IPT), under the stress- and maturation-induced promoter PSARK (Peleg et al. 2011). Root-specific expression of the Arabidopsis LHT1, amino acid permease (AtAAP) type, proline and glycine betaine (ProT) and PTR among others, under abiotic and biotic stresses, was recently reviewed by Tegeder and Rentsch (2010). These studies on the relationship between changing nutrient status under drought suggest that maintaining the nutrient status of plants may improve drought tolerance. Indeed, survival of maize plants under drought by addition of exogenous sugars revealed that the main cause for tissue mortality was not low water potential, but low carbon availability (Boyle et al. 1991). Our contention is that root proteases play an important role in nutrient uptake and mobilization through the processes discussed above, to maintain the critical nutrient balance as long as possible and thus contribute to drought tolerance. We support this hypothesis through analyzing the rice expression database for the expression of proteases in roots under drought at the critical developmental stages, as described below. Expression variation in rice root proteases under drought For total soluble protein extracts following different extraction protocols, total protease activity was apparently higher in rice roots than in shoots as seen by the extent of degradation of high-molecular-weight proteins after polyacrylamide gel electrophoresis (PAGE)mediated separation (Raorane et al. 2011). Fig. 1 shows more degradation of proteins if leaf and root tissue (L/R) proteins from one genotype were extracted together than when only leaf tissue (L) was used. The L/R samples were far more degraded than L samples to begin with and became progressively more degraded with time. That the root extract per se was responsible for the degradation of leaf proteins was demonstrated by leaf extract degradation on the addition of root extract (Raorane and Narciso, unpublished data). The addition of a protease inhibitor cocktail in the extraction buffer did not substantially alter the scenario (Raorane et al. 2011). This was true for plants at the 15-day-old seedling stage as well as at tillering stage for 12 different rice genotypes under normal non-stress conditions in hydroponics and soil-grown plants (Raorane and Narciso, unpublished data). Under Physiol. Plant. 2012 drought stress imposed on 15-day-old seedlings of the 12 rice genotypes varying for drought tolerance, preliminary results suggested a trend toward higher total root protease activity in the tolerant genotypes. This was also noticed for salinity treatment whereby a tolerant genotype such as Pokkali exhibited stronger upregulation of proteases compared to the intolerant IR29 (Miro and Raorane, unpublished data). It was not clear if this was because of more proteases in the roots of more tolerant plants. To obtain an idea on whether or not that could be the case, the rice expression database (www.ricearray.org/) was queried for changes in expression pattern of 672 proteases in roots and leaves at tillering and panicle elongation stages under control and drought conditions (Experiment GSE26280: Genome-wide temporal–spatial gene expression profiling of drought responsiveness in rice; Wang et al. 2011). The two stages were chosen for comparison because of recent trends in screening for reproductive stage drought tolerance (Serraj et al. 2009). Fig. 2 shows that in leaves and roots at the tillering or panicle development stage under drought stress, 32 proteases were upregulated twofold or more in comparison to the control plants. Of these, 11 were uniquely upregulated in roots, 5 at tillering stage and 6 at panicle elongation stage. Similarly, 15 proteases were unique to leaves, 2 at tillering stage and 13 at panicle elongation stage. Upregulation of one protease was common to roots at both stages of development M L1 L/R1 L2 L//R2 L3 L/R R3 Fig. 1. PAGE of total soluble protein of leaves (L) or leaves and roots (L/R) after incubation at 37◦ C for 10, 20 and 30 min (1, 2 and 3), respectively. The leaf extracts (L1, L2 and L3) are not affected while a progressive degradation of proteins with time is obvious in leaf and root extracts (L/R1, L/R2 and L/R3), especially in the region between the arrows. RT: 8 5 Total:: 20 RE: 12 1 1 6 2 LT: 6 To otal: 21 2 1 1 1 LE: 1 15 13 Fig. 2. Rice proteases showing more than twofold induction under drought. RT, root at tillering stage; RE, root at panicle elongation stage; LT, leaves at tillering stage; LE, leaves at panicle elongation stage. In total, 21 proteases (15 + 6) were upregulated in leaves and 20 (12 + 8) in roots. Six proteases were common to more than one box and, of those, only one was common to leaves and roots at both developmental stages. and the other 5 were upregulated in roots and leaves both at the same or different stages of development (Fig. 2). Only one protease was more than twofold upregulated in all four tissues under drought. This was an ATP-dependent La protease. The importance of such regulatory proteases in wheat under drought was described earlier (Zagdanska and Wisnievski 1998, Demirevska et al. 2008) and a distinction was made between the common vacuolar degradative proteases and the regulatory processive proteases. In our analysis, rice leaves exhibited a larger number of uniquely upregulated proteases. These comprised several aspartic proteases, including nepenthesin and the deubiquitination CPs such as a small ubiquitin-related modifier (SUMO) protease, an ovarian tumor (OTU)-like protease and four ubiquitin-like protease (Ulp) proteases. These proteases serve a regulatory function by activating the proteins that are deubiquitinated, including histones (H2B), which affects the transcriptional activation of genes such as those involved in flowering (Schmitz et al. 2009). Similarly, two serine carboxypeptidases were uniquely upregulated in leaves, which also generally help in protein maturation through post translational modifications. Other proteases were nonspecific subtilisins and CPs most likely involved in general protein degradation. In roots also, a couple of unique nepenthesins, subtilisins, serine carboxypeptidases, a Ulp protease and a CP (EP-B1) were upregulated. However, more than twofold upregulation of an M50 protease, a degradation of periplasmic protein (Deg) protease and an FtsH protease was unique to roots. In fact, these proteases were among the most highly upregulated ones in roots. The M50 proteases are a class of membrane-bound metalloproteases. Their active sites lie in transmembrane domains that participate in regulated intramembrane proteolysis (RIP). They perform site-specific proteolysis within the membranes, of other membrane-tethered (MT) proteins, to liberate the cytosolic fragment for signaling or transcription (Brown et al. 2000). RIP proteases are special in that they apparently use water to hydrolyze the peptide bond despite the hydrophobic intramembrane environment (Wolfe and Kopan 2004). RIP can drive processes ranging from differentiation, development, sterol/lipid metabolism and response to unfolded proteins. Drought conditions do induce an unfolded protein response (UPR) pathway (Zhang et al. 2008). RIP participates in this induction by liberating the cytosolic fragment of the MT transcription factor (TF) bZIP60 and bZIP28, which induce the transcription of proteinfolding chaperones, called binding proteins [(BiP); Iwata and Koizumi (2005), Liu et al. (2007)]. Also, the connection between drought, sterol levels in roots and stomatal responses has been established in Arabidopsis (Pose et al. 2009). Changes in root sterol levels can affect the sterol-regulated element binding protein (SREBP) TF. Interestingly, RIP proteases such as M50 are implicated in SREBP site 2 proteolysis (S2P) to activate the transcription factor (Rawson et al. 1997). It is not difficult to further imagine a relationship between sterol level and the UPR due to changes in sterols affecting changes in glycans, in turn affecting protein glycosylation, and thus leading to ER stress-mediated induction of UPR during drought. The upregulation of an M50 RIP protease in roots under drought thus seems logical and important. The Deg proteases are ATP-independent serine endopeptidases, which contain one or more [post synaptic density protein (PSD95), drosophila disc large tumor suppressor (DlgA) and zonula occludens-1 protein (zo1)] (PDZ) protein–protein interaction domains (Kieselbach and Funk 2003). The PDZ domain regulates the protease activity and oligomerization of Deg proteases. Deg proteases display dual function, as general proteases and as specific regulatory proteases in signal transduction or protein maturation (chaperones). This variation may be attributed to the monomer and oligomer configuration respectively. Also, Deg proteases can either have a broad substrate range or highly specific substrates. An Arabidopsis glyoxysomal Deg protease was specifically involved in the maturation of the glyoxysomal malate dehydrogenase (Helm et al. 2007). Deg proteases have not been characterized in rice and Physiol. Plant. 2012 root-specific or drought-inducible Deg proteases are not known. However, a Deg protease was earlier shown to be relevant to protein quality control in chloroplasts (Huesgen et al. 2009) through a role in repairing UV-B damage of PSII (Nixon et al. 2010). In this particular function the Deg proteases had a complementary role with the FtsH proteases, which are ATP-dependent, Znmetalloproteases. Both Deg and FtsH proteases are rather known as plastidial proteases in leaves and not in root. An FtsH protease was characterized to be responsible for the variegated phenotype through affecting chloroplast development (Pogson and Albrecht 2011). Therefore, root-specific upregulation of rice Deg and FtsH proteases under drought warrants attention. It is likely that their function in degrading unassembled membrane proteins and/or oxidatively damaged proteins makes them relevant in drought as part of the UPR pathway similar to the M50 and rhomboid proteases. It is noteworthy that proteases highly and uniquely upregulated in rice roots under drought, although belonging to different classes, may be concerned with the RIP and UPR pathways both of which can either affect the C–N pool under stress by internal proteolysis and/or by facilitating external nutrient uptake as discussed above or affect protein processing and maturation for signaling and transcription. Additionally, our in silico analysis revealed some proteases which, although not twofold upregulated under drought, were specifically induced in response to drought in both leaves and roots at all developmental stages assessed by Wang et al. (2011) (Fig. 3, Table 1). Most of these were similar to proteases discussed above for example, a SUMO protease, a Deg protease, an SCP, an OTU-like CP, and an M16 domain containing Zinc peptidase similar to the M50 peptidase. However, there were four unique proteases, a rhomboid (serine) protease, an AMSH-like protease, a signal peptide peptidase and a mitochondrial processing peptidase. Rhomboid proteases are also a part of the RIP arsenal and are substrate-specific (Kanaoka et al. 2005). The potential role of these proteases under drought assumes more importance with the knowledge that plants can endocytose large organic molecules and microbes (Paungfoo-Lonhienne et al. 2010), which invokes mucilage-mediated attraction and receptor-mediated endocytosis of molecules/microbes. Both of these can be imagined to be affected by root RIP proteases upregulated under drought. Interestingly, the AMSH protease is closely associated with clathrin-mediated endosomal transport of nutrients through symplastic or apoplastic movements from one part of the plant to another. The other two peptidases are also processing peptidases that enable protein trafficking to the correct subcellular compartments. Physiol. Plant. 2012 Fig. 3. Drought-induced upregulation of a rhomboid protease designated by the oligo-array element, in leaves and roots at different developmental stages. An arbitrary comparative estimate of an increase in protease activity contributed by the twofold or more upregulated proteases in roots and leaves (Fig. 2) was obtained by the sum of fold increases in roots (20 proteases upregulated 52.57-fold) and leaves (21 proteases upregulated 54.69-fold). The result that a higher number of proteases were upregulated and to a larger extent in leaves than in roots was contrary to the observation of higher and more potent root protease activity. This suggests that root proteases may be highly degradative and/or processive. The FtsH protease from Synechocystis is indeed highly processive (Komenda et al. 2007) and such an attribute is likely retained in rice root proteases. Root proteases and crop improvement Increasing biomass and seed yield is mostly the ultimate goal of a large body of agricultural research. However, plant growth and productivity are directly and strongly affected by edaphic and other environmental factors, most of which are perceived by, responded to and effected on roots. Yet, root responses at the physiological and/or molecular levels are poorly understood. Although it is becoming increasingly appreciated that optimal plant growth and productivity can best be achieved by Table 1. Proteases specifically induced in response to drought in both leaves and roots at all developmental stages assessed under experiment GSE26280 by Wang et al. (2011) Array element ID Os.10283.1.S1_at Os.11236.1.S1_at Os.13615.1.S1_at Os.4124.1.S1_a_at Os.4146.1.S1_at Os.46745.1.S1_at Os.53632.1.S1_at Os.6594.1.S1_at Os.8776.1.S1_at OsAffx.24819.1.S1_at OsAffx.24834.1.S1_x_at Gene ID Gene name LOC_Os04g01240 LOC_Os02g12650 LOC_Os01g47262 LOC_Os01g51390 LOC_Os01g25370 LOC_Os02g57710 LOC_Os05g13370 LOC_Os11g24510 LOC_Os03g64219 LOC_Os02g50880 LOC_Os02g52390 Serine-type peptidase Puromycin-sensitive aminopeptidase AMSH-like protease, putative, expressed Mitochondrial-processing peptidase SUMO protease Signal peptide peptidase-like 2B OsRhmbd12–putative rhomboid protease OsSCP56–serine carboxypeptidase OTU-like cysteine protease family protein OsDegp3–putative Deg protease M16 domain containing zinc peptidase better understanding and leveraging the role of roots, several issues are emerging in how best to strengthen the desirable traits of roots while avoiding their undesirable traits for overall crop improvement (Herder et al. 2010). Recent studies have highlighted unconventional sources and routes of nutrient uptake by roots, while strong links between drought tolerance and nutrient status have also been highlighted above. These studies suggest a central role for root proteases not only in nutrient uptake, but also in water uptake through processing aquaporins for their role in maintaining plant water potential (Sade et al. 2009, Maurel et al. Fig. 4. A schematic model for the role of proteases in proteinaceous nutrition uptake through different modes by root cells at the soil interface. Left side (beige) represents the cell cytoplasm, cell membrane ad cell wall, while the right side represents the soil. Plant proteases (brown globules) and/or microbial proteases (yellow globules) act on bacteria (red rods) and proteins (green zigzags) in the soil to generate peptides (small green rods) and/or amino acids (green AA). Similarly such peptides and AAs are generated within the plant cell by the action of endogenous plant proteases on endogenous proteins. Additionally, root exudates (evaginated pustule at cell–soil interface) may comprise plant proteins and peptides further degraded at the rhizoplane or in the soil into peptides and AAs by exuded plant proteases or microbial proteases. It is known that peptides and AAs serve as sources of carbon and nitrogen through further breakdown within the plant cell. Peptide and AA uptake is well known. However, under drought conditions, apparently intact proteins and complete bacterial and yeast cells are also taken up by the root cells through the process of endocytosis (invaginated pustule). Under well-watered conditions root exudates and other process may facilitate most part of the breakdown through microbial action in the soil, thus enabling re-uptake of simpler peptides and AAs. However, under drought conditions most breakdown and uptake may happen at the rhizoplane or within the cells, facilitated by endocytosis of larger proteins and cells. Physiol. Plant. 2012 2010). Yet, very limited knowledge exists on the identity and function of root proteases under normal or stress conditions. A better understanding of root proteases will lead to designing robust strategies for crop improvement by using proteases as gene-based markers for genotyping and population surveys or through transgenic approaches. Regulatory proteases can play an important part in crop improvement because they, like transcription factors and kinases, can affect multiple cascades through spatio-temporalspecific protein processing, leading perhaps to a burst of response without necessarily involving transcription for a part of the response. Although this article concentrated on establishing the links between root proteases and water and nutrient uptake and mobilization during drought stress, proteases functional in root growth, branching and surface may have an effect on aerial tissues. Root apical and lateral meristem activity and root vascular development are controlled by Cle peptides, which are the result of specific processing proteases (Ni et al. 2010). Processing of proteins facilitating symplastic movement or xylem to phloem transporters may also be achieved by proteases. Studies on proteases in general and root proteases in particular are also important to differentiate between desirable and undesirable proteases under specific conditions. An understanding of root function for water and nutrient uptake and mobilization, which is inclusive of studies on root proteases, would fast-track our physiological and molecular understanding of stress tolerance. This would lead to informed strategies for crop improvement under conditions of adverse climate and shrinking agricultural lands and help meet the goals of adequate food and nutrition for the increasing population. Conclusions and perspectives Root proteases may have a role in water and nutrient uptake and mobilization, the quality and quantity of rhizodeposition, microbial population modulation and root function as a source and sink for nutrients. Increasing evidence suggests that root proteases may be an important aspect of root development, differentiation and stress signaling. Various structural and functional classes of proteases make it difficult to generalize the role of proteases in a process, for example, drought-tolerant plants may exhibit upregulation of some proteases and downregulation of others. Tissue-specific protease isozymes also exhibit functional diversity, for example, chloroplast proteases such as Deg, Clp, FtsH, etc. are upregulated in roots, where they may not at all be connected with processing photosynthesis-related proteins. Such inherent Physiol. Plant. 2012 diversity and the difficulty of working with roots under natural conditions pose a challenge in characterizing this important class of proteins. It is certain, however, that making substantial headway in understanding and affecting above-ground tissues toward crop improvement will critically rest on our understanding of roots. In that regard, root proteases will be only a part of the sum, but a very important part, since their capacity to regulate cascades of reactions places them at par with other protein modification systems such as kinases. Recent literature clearly makes a case for the importance of root proteases in drought tolerance. Fig. 4 presents a schematic version of the role of proteases in nutrient uptake especially under drought conditions. Drought is a major bottleneck in food security at present and even more so in the climate change-driven future scenarios of agriculture. It is clear that addressing drought tolerance only in terms of water uptake, movement and use efficiency will be only half the understanding on drought tolerance because nutrient status imbalances under drought apparently affect the plant sooner than the lack of water. Root proteases can positively affect nutrient and water status more than any other class of proteins under drought where anabolic processes in the above-ground source tissues start suffering even before critically low levels of water are reached. Research on drought tolerance must therefore put equal if not more emphasis on the physiology and molecular biology of the nutrient status of plants under drought and as a corollary on proteases in general and root proteases in particular. Acknowledgements – We thank Drs Akshaya Kumar Biswal, Abdelbagi Ismail, Roland Buresh and Stephan Haefele of IRRI for critical comments and suggestions on improving the manuscript. We thank Dr Bill Hardy of IRRI for improving the manuscript through meticulous editorial corrections. References Adamczyk B, Godlewski M, Smolandera A, Kitunen V (2009) Degradation of proteins by enzymes exuded by Allium porrum roots – a potentially important strategy for acquiring organic nitrogen by plants. Plant Physiol Biochem 47: 919–925 Adamczyk B, Godlewski M, Zimny J, Zimny A (2008) Wheat (Triticumaestivum) seedlings secrete proteases from the roots and, after protein addition, grow well on medium without inorganic nitrogen. Plant Biol 10: 718–724 Atkins CA (2000) Biochemical aspects of assimilate transfers along the phloem path: N-solutes in lupins. Aust J Plant Physiol 27: 531–537 Baek KH, Choi D (2008) Roles of plant proteases in pathogen defense. Plant Pathol J 24: 367–374 Bathellier C, Tcherkez G, Mauve C, Bligny R, Gout E, Ghashghaie J (2009) On the resilience of nitrogen assimilation by intact roots under starvation, as revealed by isotopic and metabolomic techniques. Rapid Commun Mass Spectrom 23: 2847–2856 Bick JA, Neelam A, Hall JL, Williams LE (1998) Amino acid carriers of Ricinus communis expressed during seedling development: molecular cloning and expression analysis of two putative amino acid transporters, RcAAP1 and RcAAP2. Plant Mol Biol 36: 377–385 Bingham IJ, Rees RM (2008) Senescence and N release from clover roots following permanent excision of the shoot. Plant Soil 303: 229–240 Boyle MG, Boyer JS, Morgan PW (1991) Stem infusion of liquid culture medium prevents reproductive failure of maize at low water potential. Crop Sci 31: 1246–1252 Brouquisse R, Evrard A, Rolin D, Raymond P, Roby C (2001) Regulation of protein degradation and protease expression by mannose in maize root tips: Pi sequestration by mannose may hinder the study of its signaling properties. Plant Physiol 125: 1485–1498 Brouquisse R, Rolin D, Cortes S, Gaudillere M, Evrard A, Roby C (2007) A metabolic study of the regulation of proteolysis by sugars in maize root tips: effects of glycerol and dihydroxyaceton. Planta 225: 693–709 Brown MS, Ye J, Rawson RB, Goldstein JL (2000) Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100: 391–398 Buresh RJ, Reddy KR, van Kessel C (2008) Nitrogen transformations in submerged soils. In: Schepers JS, Raun WR (eds) Nitrogen in Agricultural Systems. Agronomy Monograph 49. ASA, CSSA, and SSSA, Madison, WI, pp 401–436 Casamitjana-Martinez E, Hofhuis HF, Xu J, Liu CM, Heidstra R, Scheres B (2003) Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr Biol 13: 1435–1441 Chen P, Sneller CH, Purcell LC, Sinclair TR, King TR, Ishibashi CAT (2007) Registration of soybean germplasm lines R01-416F and R01-581F for improved yield and nitrogen fixation under drought stress. J Plant Regist 1: 167–168 Chen YF, Wang Y, Wu WH (2008) Membrane transporters for nitrogen, phosphate and potassium uptake in plants. J Integr Plant Biol 50: 835–848 Chevalier C, Bourgeois E, Pradet A, Raymond P (1995) Molecular cloning and characterization of six cDNAs expressed during glucose starvation in excised maize (Zea mays L.) root tips. Plant Mol Biol 28: 473–485 Christmann A, Weiler EW, Steudle E, Grill E (2007) A hydraulic signal in root-to-shoot signalling of water shortage. Plant J 52: 167–174 Combier JP, Vernie T, de Billy F, El Yahyaoui F, Mathis R, Gamas P (2007) The Mt MMPL1 early nodulin is a novel member of the matrix metalloendoproteinase family with a role in Medicago truncatula infection by Sinorhizobium meliloti. Plant Physiol 144: 703–716 De-la-Pena C, Vivanco JM (2010) Root-microbe interactions: the importance of protein secretion. Curr Proteomics 7: 265–274 Demirevska K, Simova-Stoilova L, Vassileva V, VasevaI Grigorova B, Feller U (2008) Drought-induced leaf protein alterations in sensitive and tolerant wheat varieties. Gen Appl Plant Physiol 34: 79–102 Devaux C, Baldet P, Joubes J, Dieuaide-Noubhani M, Just D, Chevalier C, Raymond P (2003) Physiological, biochemical and molecular analysis of sugar-starvation responses in tomato roots. J Exp Bot 54: 1143–1151 Dieuaide-Noubhani M, Canioni P, Raymond P (1997) Sugar-starvation-induced changes of carbon metabolism in excised maize root tips. Plant Physiol 115: 1505–1513 Duan YF, Zhang WS, Li B, Wang YN, Li K, Sodmergen Han C, Zhang Y, Li X (2010) An endoplasmic reticulum response pathway mediates programmed cell death of root tip induced by water stress in Arabidopsis. New Phytol 186: 681–695 Farooq M, Wahid A, Kobayashi N, Fujita D, Basra SMA (2009) Plant drought stress: effects, mechanisms and management. Agron Sustain Dev 29: 185–212 Gaufichon L, Reisdorf-Cren M, Rothstein SJ, Chardon F, Suzuki A (2010) Biological functions of asparagine synthetase in plants. Plant Sci 179: 141–153 Godlewski M, Adamczyk B (2007) The ability of plants to secrete proteases by roots. Plant Physiol Biochem 45: 657–664 Gonzalez-Dugo V, Durand JL, Gastal F (2010) Water deficit and nitrogen nutrition of crops. Agron Sustain Dev 30: 529–544 Guo SW, Chen G, Zhou Y, Shen QR (2007) Ammonium nutrition increases photosynthesis rate under water stress at early development stage of rice (Oryza sativa L.). Plant Soil 296: 115–124 Hartmann A, Schmid M, van Tuinen D, Berg G (2009) Plant-driven selection of microbes. Plant Soil 321: 235–257 Helm M, Lück C, Prestele J, Hierl G, Huesgen PF, Fröhlich T, Arnold GJ, Adamska I, Görg A, Lottspeich F, Gietl C (2007) Dual specificities of the glyoxysomal/peroxisomal processing protease Deg15 in higher plants. Proc Natl Acad Sci USA 104: 11501–11506 Herder GD, Isterdael GV, Beeckman T, Smet ID (2010) The roots of a new green revolution. Trends Plant Sci 15: 600–607 Physiol. Plant. 2012 Hieng B, Ugrinović K, Sustar-Vozlic J, Kidric M (2004) Different classes of proteases are involved in the response to drought of Phaseolus vulgaris L. cultivars differing in sensitivity. J Plant Physiol 161: 519–530 Hill PW, Quilliam RS, DeLuca TH, Farrar J, Farrell M, et al. (2011) Acquisition and Assimilation of Nitrogen as Peptide-Bound and D-Enantiomers of Amino Acids by Wheat. PLoS ONE 6(4): e19220. doi:10.1371/journal. pone.0019220 Hirner A, Ladwig F, Stransky H, Okumoto S, Keinath M, Harms A, Frommer WB, Koch W (2006) Arabidopsis LHT1 is a high-affinity transporter for cellular amino acid uptake in both root epidermis and leaf mesophyll. Plant Cell 18: 1931–1946 Hodge A, Berta G, Doussan C, Merchan F, Crespi M (2009) Plant root growth, architecture and function. Plant Soil 321: 153–187 Hu Y, Schmidhalter U (2005) Drought and salinity: a comparison of their effects on the mineral nutrition of plants. J Plant Nutr Soil Sci 168: 541–549 Huesgen PF, Schuhmann H, Adamsk I (2009) Deg/HtrA proteases as components of a network for photosystem II quality control in chloroplasts and cyanobacteria. Res Microbiol 160: 726–732 Iwata Y, Koizumi N (2005) An Arabidopsis transcript ion factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc Natl Acad Sci USA 102: 5280–5285 Jones DL, Nguyen C, Finlay RD (2009) Carbon flow in the rhizosphere: carbon trading at the soil-root interface. Plant Soil 321: 5–33 Jones JT, Mullet JE (1995) A salt-inducible and dehydrationinducible pea gene, Cyp15a, encodes a cell-wall protein with sequence similarity to cysteine proteases. Plant Mol Biol 28: 1055–1065 Kanaoka M, Urban S, Freeman M, Okada K (2005) An Arabidopsis rhomboid homolog is an intramembrane protease in plants. FEBS Lett 579: 5723–5728 Khanna-Chopra R, Srivalli B, Ahlawat YS (1999) Drought induces many forms of cysteine proteases not observed during natural senescence. Biochem Biophys Res Commun 255: 324–327 Kieselbach T, Funk C (2003) The family of Deg/HtrA proteases: from Escherichia coli to Arabidopsis. Physiol Plant 119: 337–346 Koizumi M, Yamaguchi-Shinozaki K, Tsuji H, Shinozaki K (1993) Structure and expression of two genes that encode distinct drought-inducible cysteine proteinases in Arabidopsis thaliana. Gene 129: 175–182 Komenda J, Tichy M, Prasil O, Knoppova J, Kuvikova S, de Vries R, Nixon PJ (2007) The exposed N-terminal tail of the D1 subunit is required for rapid D1 degradation during photosystem II repair in Synechocystissp PCC 6803. Plant Cell 19: 2839–2854 Physiol. Plant. 2012 Kraiser T, Gras DE, Gutierrez AG, Gonzalez B, Gutierrez RA (2011) A holistic view of nitrogen acquisition in plants. J Exp Bot 62: 1455–1466 Krcek M, Slamka P, Olsovska K, Brestic M, Bencikova M (2008) Reduction of drought stress effect in spring barley (Hordeumvulgare L.) by nitrogen fertilization. Plant Soil Environ 54: 7–13 Kuroda H, Maliga P (2003) The plastid clpP1 protease gene is essential for plant development. Nature 425: 86–89 Leborgne-Castel N, Adam T, Bouhidel K (2010) Endocytosis in plant-microbe interactions. Protoplasma 247: 177–193 Lesuffleur F, Paynel F, Bataille MP, Le Deunff E, Cliquet JB (2007) Root amino acid exudation: measurement of high efflux rates of glycine and serine from six different plant species. Plant Soil 294: 235–246 Li YX, Zhou L, Li YG, Chen DS, Tan XJ, Lei L, Zhou JC (2008) A nodule-specific plant cysteine proteinase, AsNODF32, is involved in nodule senescence and nitrogen fixation activity of the green manure legume Astragalus sinicus. New Phytol 180: 185–192 Liu JX, Srivastava R, Che P, Howell SH (2007) An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane associated transcription factor, bZIP28. Plant Cell 19: 4111–4119 Liu X, Yu F, Rodermel S (2010) Arabidopsis chloroplast FtsH, var2 and suppressors of var2 leaf variegation: a review. J Integr Plant Biol 52: 750–761 Lopez-Bucio J, Cruz-Ramirez A, Herrera-Estrella L (2003) The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol 6: 280–287 Lubkowitz M (2011) The oligopeptide transporters: a small gene family with a diverse group of substrates and functions? Mol Plant 4: 407–415. Maurel C, Simonneau T, Sutka M (2010) The significance of roots as hydraulic rheostats. J Exp Bot 61: 3191–3198 McDowell NG (2011) Mechanisms linking drought, hydraulics, carbon metabolism, and vegetation mortality. Plant Physiol 155: 1051–1059 McDowell NG, White S, Pockman WT (2008) Transpiration and stomatal conductance across a steep climate gradient in the southern Rocky Mountains. Ecohydrology 1: 193–204 Murray JD (2011) Invasion by invitation: rhizobial infection in legumes. Mol Plant Microbe Interact 24: 631–639 Nasholm T, Kielland K, Ganeteg U (2009) Uptake of organic nitrogen by plants. New Phytol 182: 31–48 Ni J, Guo Y, Jin H, Hartsell J, Clark SE (2010) Characterization of a CLE processing activity. Plant Mol Biol 75: 67–75 Nixon PJ, Michoux F, Yu J, Boehm M, Komenda J (2010) Recent advances in understanding the assembly and repair of photosystem II. Ann Bot 106: 1–16 Okumoto S, Pilot G (2011) Amino acid export in plants: a missing link in nitrogen cycling. Mol Plant 4: 453–463. Panigrahy M, Rao DN, Sarla N (2009) Molecular mechanisms in response to phosphate starvation in rice. Biotechnol Adv 27: 389–397 Paterson E, Thornton B, Sim A, Pratt S (2003) Effects of defoliation and atmospheric CO2 depletion on nitrate acquisition, and exudation of organic compounds by roots of Festuca rubra. Plant Soil 250: 293–305 Paungfoo-Lonhienne C, Lonhienne TGA, Rentsch D, Robinson N, Christie M, Webb RI, Gamage HK, Carroll BJ, Schenk PM, Schmidt S (2008) Plants can use protein as a nitrogen source without assistance from other organisms. Proc Natl Acad Sci USA 105: 4524–4529 Paungfoo-Lonhienne C, Rentsch D, Robatzek S, Webb RI, Sagulenko E, Nasholm T, Schmidt S, Lonhienne TGA (2010) Turning the table: plants consume microbes as a source of nutrients. PLoS One 5: e11915 Peleg Z, Reguera M, Tumimbang E, Walia H, Blumwald E (2011) Cytokinin-mediated source/sink modifications improve drought tolerance and increase grain yield in rice under water-stress. Plant Biotechnol J 9: 747–758 Persson J, Nasholm T (2002) Regulation of amino acid uptake in conifers by exogenous and endogenous nitrogen. Planta 215: 639–644 Pimratch S, Jogloy S, Vorasoot N, Toomsan B, Patanothai A, Holbrook CC (2008) Relationship between biomass production and nitrogen fixation under drought-stress conditions in peanut genotypes with different levels of drought resistance. J Agron Crop Sci 194: 15–25 Pinheiro C, Chaves MM (2011) Photosynthesis and drought: can we make metabolic connections from available data? J Exp Bot 62: 869–882 Pogson BJ, Albrecht V (2011) Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiol 155: 1545–1551 Pose D, Castanedo I, Borsani O, Nieto B, Rosado A, Taconnat L, Ferrer A, Dolan L, Valpuesta V, Botella MA (2009) Identification of the Arabidopsis dry2/sqe1-5 mutant reveals a central role for sterols in drought tolerance and regulation of reactive oxygen species. Plant J 59: 63–76 Raorane M, Narciso JO, Kohli A (2011) Total soluble protein extraction for improved proteomic analysis of transgenic plants. In: Twyman RM, Christou P (eds) Methods in Molecular Biology: Multigene Engineering in Plants. Springer-Verlag, New York. (in press) Rawson R, Zelenski N, Nijhawan D, Ye J, Sakai J, Hasan M, Chang T, Brown M, Goldstein J (1997) Complementation cloning of SP2, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol Cell 1: 47–57 Rose TL, Bonneau L, Der C, Marty-Mazars D, Marty F (2006) Starvation-induced expression of autophagy-related genes in Arabidopsis. Biol Cell 98: 53–67 Rosengren-Brinck U, Nihlgard B (1995) Effects of nutritional status on the drought resistance in Norway spruce. Water Air Soil Pollut 85: 1739–1744 Rost TL (2011) The organization of roots of dicotyledonous plants and the positions of control points. Ann Bot 107: 1213–1222 Rouached H, Secco D, Arpat AB (2009) Getting the most sulfate from soil: regulation of sulfate uptake transporters in Arabidopsis. J Plant Physiol 166: 893–902 Roy-Macauley H, Zuily-Fodil Y, Kidric M, Pham Thi AT, Vieira da Silva J (1992) Effect of drought stress on proteolytic activities in Phaseolus and Vigna leaves from sensitive and resistant plants. Physiol Plant 85: 90–96 Sade N, Vinocur BJ, Diber A, Shatil A, Ronen G, Nissan H, Wallach R, Karchi H, Moshelion M (2009) Improving plant stress tolerance and yield production: is the tonoplast aquaporin SlTIP2;2 a key to isohydric to anisohydric conversion? New Phytol 181: 651–661 Sardans J, Penuelas J (2005) Drought decreases soil enzyme activity in a Mediterranean Quercus ilex L. forest. Soil Biol Biochem 37: 455–461 Schaller A (2004) A cut above the rest: the regulatory function of plant proteases. Planta 220: 183–197 Schmitz RJ, Tamada Y, Doyle MR, Zhang X, Amasino RM (2009) Histone H2B deubiquitination is required for transcriptional activation of FLOWERING LOCUS C and for proper control of flowering in Arabidopsis. Plant Physiol 149: 1196–1204 Schobert C, Komor E (1987) Amino acid uptake by Ricinus communis roots: characterization and physiological significance. Plant Cell Environ 10: 493–500 Schobert C, Komor E (1990) Transfer of amino acids and nitrate from the roots into the xylem of Ricinus communis seedlings. Planta 181: 85–90 Serraj R, Kumar A, McNally KL, Slamet-Loedin I, Bruskiewich R, Mauleon R, Cairns J, Hijmans RJ (2009) Improvement of drought resistance in rice. Adv Agron 103: 41–99 Shi K, Ding XT, Dong DK, Zhou YH, Yu JQ (2008) Root restriction-induced limitation to photosynthesis in tomato (Lycopersicon esculentum Mill.) leaves. Sci Hortic 17: 197–202 Simova-Stoilova L, Vaseva I, Grigorova B, Demirevska K, Feller U (2010) Proteolytic activity and cysteine protease expression in wheat leaves under severe soil drought and recovery. Plant Physiol Biochem 48: 200–206 Simova-Stoilova L, Vassileva V, Petrova T, Tsenov N, Demirevska K, Feller U (2006) Proteolytic activity in wheat leaves during drought stress and recovery. Gen Appl Plant Physiol 32: 91–100 Physiol. Plant. 2012 Simpson K (2011) Transport of nitrogen in plants. PhD Dissertation, University of Sheffield, Sheffield. www.shef.ac.uk/aps/mbiolsci/kelly/dissertation.pdf (Accessed 28 July 2011) Smith AM, Stitt M (2007) Coordination of carbon supply and plant growth. Plant Cell Environ 30: 1126–1149 Suralta RR (2010) Plastic root system development responses to drought-enhanced nitrogen uptake during progressive soil drying conditions in rice. Philipp Agric Sci 93: 458–462 Szczerba MW, Britto DT, Kronzucker HJ (2009) K+ transport in plants: physiology and molecular biology. J Plant Physiol 166: 447–466 Tegeder M, Rentsch D (2010) Uptake and partitioning of amino acids and peptides. Mol Plant 3: 997–1011 Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD (2005) Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol 138: 2097–2110 Thornton B, Osborne SM, Paterson E, Cash P (2007) A proteomic and targeted metabolomic approach to investigate change in Lolium perenne roots when challenged with glycine. J Exp Bot 58: 1581–1590 Vágnerová K, Macura J (1974) Relationships between plant roots, proteolytic organisms and activity of protease. Folia Microbiol 19: 525–535 Van der Hoorn R (2008) Plant proteases: from phenotypes to molecular mechanisms. Annu Rev Plant Biol 59: 191–223 Wang D, Pan Y, Zhao X, Zhu L, Fu B, Li Z (2011) Genome-wide temporal-spatial gene expression profiling of drought responsiveness in rice. BMC Genom 12: 149 Webb KJ, Jensen EF, Heywood S, Morris SM, Linton PE, Hooker JE (2010) Gene expression and nitrogen loss in senescing root systems of red clover (Trifolium pratense). J Agric Sci 148: 579–591 Edited by E. Pesquet Physiol. Plant. 2012 Wisniewski K, Zagdanska B (2001) Genotype-dependent proteolytic response of spring wheat to water deficiency. J Exp Bot 52: 1455–1463 Wolfe MS, Kopan R (2004) Intramembrane proteolysis: theme and variations. Science 305: 1119–1123 Woltering EJ (2004) Death proteases come alive. Trends Plant Sci 9: 469–472 Xia YJ (2004) Proteases in pathogenesis and plant defence. Cell Microbiol 6: 905–913 Yang H, Bogner M, Stierhof YD, Ludewig U (2010) H+ -Independent glutamine transport in plant root tips. PLoS One 5: e8917 Yokota K, Hayashi M (2011) Function and evolution of nodulation genes in legumes. Cell Mol Life Sci 68: 1341–1351 Zagdanska B, Wisniewski K (1996) Endoproteinase activities in wheat leaves upon water deficit. Acta Biochim Pol 43: 515–520 Zagdanska B, Wisniewski K (1998) ATP-dependent proteolysis contributes to the acclimation-induced drought resistance in spring wheat. Acta Physiol Plant 20: 41–48 Zang QW, Wang CX, Li XY, Guo ZA, Jjing RL, Zhao J, Chang XP (2010) Isolation and characterization of a gene encoding a polyethylene glycol-induced cysteine protease in common wheat. J Biosci 35: 379–388 Zhang H, Ohyama K, Boudet J, Chen Z, Yang J, Zhang M, Muranaka T, Maurel C, Zhu JK, Gong Z (2008) Dolichol biosynthesis and its effects on the unfolded protein response and abiotic stress resistance in Arabidopsis. Plant Cell 20: 1879–1898 Zhang L, Tan Q, Lee R, Trethewy A, Lee YH, Tegeder M (2010) Altered xylem-phloem transfer of amino acids affects metabolism and leads to increased seed yield and oil content in Arabidopsis. Plant Cell 22: 3603–3620