* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download talk Breman 2013 cichlid fish bol5 cover
Habitat conservation wikipedia , lookup
Unified neutral theory of biodiversity wikipedia , lookup
Occupancy–abundance relationship wikipedia , lookup
Biodiversity action plan wikipedia , lookup
Molecular ecology wikipedia , lookup
Latitudinal gradients in species diversity wikipedia , lookup
Introduced species wikipedia , lookup
Fauna of Africa wikipedia , lookup
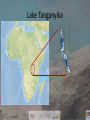
DNA barcoding LT cichlids of the rocky shores Breman FC1, Van Steenberge M2, Jordaens K1 & Snoeks J2 1 Royal Museum for Central Africa 2 Royal Museum for Central Africa and Catholic University of Leuven • • DNA barcoding of LT cichlids of the rocky shores Introduction to the topic Methods – – – – – – • • Collection Specimens Methods • Specimens • Data and library setup • Sequence based • Species based Analysis • Sequence based identification • Species based identification Results Tropheus duboisi • Sequence based • Species based • Species complexes • Example of a complex and a newly described species Testing OTU´s Discussion Questions? Introduction • Fish DNA barcoding successful so far • Most species can be identified • Different methods can be used Introduction • Habitat – rocky shores of Lake Tanganyika – Alkaline environment (average pH 8,4) – Highly diverse habitats – Highly specialized • Breeding strategies – Mouth brooders – Shell brooders – Substrate brooders • Feeding strategies – Predation – Algae scraping Introduction • 200+ recognized species of cichlids – Dozens remain to be described – >95% endemic • Model organisms for evolution and speciation • Economic importance with aquarium enthusiasts • 75 non cichlid fishes are also present in LT Lake Tanganyika Collection sites • 676 km from N-S and average 50 km across • Average depth 570m (max 1470m) • 4 countries – – – – Tanzania Congo Burundi Zambia Collection sites • 15 sites • 3 expeditions (1992, 1995, 2010) • 1000s of specimens Lobochilotus labiatus Lepdiolamprologus elongatus Tropheus brichardi Gnatohochromis pfefferi Methods Specimens • Covering 11 tribii and 37 genera • Library A) 78 (98 with singletons) OTUs • Library B) 70 (91 with singletons) OTUs • Library C) 52 (66 with singletons) OTUs (11 complexes) Methods Data setup • Three groups – A) all taxonomic, behavioural and distributional knowledge (published and unpublished) were used to assign a name to a specimen – B) only currently recognized species used for assigning a name to a specimen – C) groups with known difficulties in evolutionary history (hybridization, incomplete lineage sorting) and taxonomy are grouped in species clusters Methods Sequence based – BM/BCM method – Software compares each sequence to all the others and the chosen threshold for the dataset – Returns a statement on the sequence/specimen with regard to threshold and presence of same/related species – Returns a success percentage in terms of sequences – Influenced by dataset properties Methods Species based – Uses the sequences assigned in the sequence based method, but now classified according to threshold per species – Species identifiable or not – Returns success percentage in terms of species Analysis Species based BM/BCM • Categories – True Negative (TN) Best match above threshold and allospecific – True Positive (TP) Best match is same species below threshold – False Positive (FP) Closest match is below threshold but is an allospecific – False Negative (FN) Intraspecific distances are above threshold and conspecific FP Threshold FN • Fixed or based on dataset, in our case threshold was determined via an R script • Obtained via two curves of intra and inter specific distances • The optimum is then chosen as threshold Analysis Species based using a NJ tree • Using a NJ tree • Counting species in distinct clusters Results Sequence based (A)morphospecies 1.91% 398 (B)accepted names 2.37% 398 (C) species complexes 2.20% 398 Correct id's according to "Best Match": Ambiguous according to "Best Match": Incorrect id's according to "Best Match": 276 (69.34%) 80 (20.1%) 42 (10.55%) 320 (80.4%) 36 (9.04%) 42 (10.55%) 352 (88.44%) 21 (5.27%) 25 (6.28%) Correct id's according to "Best Close Match": Ambiguous according to "Best Close Match": 274 (68.84%) 80 (20.1%) 318 (79.89%) 36 (9.04%) 349 (87.68%) 21 (5.27%) 31 (7.78%) 34 (8.54%) 17 (4.27%) 13 (3.26%) 10 (2.51%) 11 (2.76%) optimal threshold Sequences: Incorrect id's according to "Best Close Match": Sequences without any match closer than threshold Results Species based Library A B C # species 78 70 52 TP 55 46 40 A B C Relative ID error 0.20 0.31 0.07 FP 14 21 3 Precision 0.80 0.69 0.93 TN 5 2 6 FN 4 1 3 Overall ID error 0.23 0.31 0.12 -Marginal improvement when species complexes in success %, -but an increase in accuracy and precision percentage ID success 0.71 0.59 0.51 Accuracy 0.77 0.69 0.88 % success NJ 0.74 0.74 0.81 Results Species complexes Results Species complex and new species • Examples of species complexes with unresolved clusters • Eretmodus cyanostictus specimens have recently been described as Eretmodus marksmithi Results • Identification of putative new species with DNA barcodes intrasp. dist. < intersp putative species based on # sequences dist. to nearest other dist. to nearest expert opinion in dataset species (y/n) other species Chalinochromis spbifrenatus 1 NA 2.33 Ectodus cfdescampsi 3 y 1.7 Neolamprologus cfpetricola 1 NA 0.61 Neolamprologus speseki 3 y 1.07 Petrochromis cfmacrognathus 2 y 0.15 Petrochromis spephippiumsouth 2 y 1.39 Petrochromis sppolyodonelongate 3 y 0.76 Petrochromis sppolyodonhigh 2 n 0.15 Tropheus cfannectens 8 y 1.54 Tropheus ikola 3 n 0 Tropheus mpimbwe 51 n 0 average generic distance +/- se 1.84 +/- 0.42 1.45 +/- 0.33 5.70 +/- 0.55 5.70 +/- 0.55 2.45 +/- 0.36 putative species y/n (difference > generic average +/- se) y n n n n 2.45 +/- 0.36 n 2.45 +/- 0.36 2.45 +/- 0.36 1.55 +/- 0.26 1.55 +/- 0.26 1.55 +/- 0.26 n n n n n Discussion • LT cichlids well studied – Genetically and morphologically • They are examples of ongoing speciation – mtDNA evolution is slower than morphological evolution • Hybridisation is common Discussion • Incomplete taxonomy • Unbalanced dataset, influence of sequence composition • Success percentages not high compared to other fish groups, however still ok for a complex group such as this. • Distance method not very useful for detecting potential new species • Single sequences cannot be evaluated with either method Thank you for your attention 感谢您的关注 Questions? Foto credits: Royal Museum for Central Africa Maarten Van Steenberge Dimitri Geelhand de Merxem